Unraveling the protein map of cell’s powerhouse
Mitochondria, the so-called powerhouse of the cells, are responsible for the energy supply of the organism and fulfill functions in metabolic and signaling processes. Researchers at the University Hospital Bonn and the University of Freiburg have gained systematic insight into the organization of proteins in mitochondria. The protein map of mitochondria represents an important basis for further functional characterization of the powerhouse of cell and thus provide implications for diseases. The study has now been published in the renowned scientific journal Nature.
Mitochondria are among the most important cell compartments. They are delimited organelles surrounded by a double membrane. Mitochondria are considered the powerhouse of the cell, as they produce the majority of the energy required for all cellular processes. In addition, they take over many other functions in metabolism and provide a signaling surface for inflammatory processes and programmed cell death. Defects in mitochondria lead to numerous diseases, especially of the nervous system.
Therefore, the molecular understanding of mitochondrial processes is of highest relevance for basic medical research. The molecular workers in the cell are usually proteins. Mitochondria can contain around 1,000 or more different proteins. To execute functions, several of these molecules often work together and form a protein machine, also called a protein complex. Proteins also interact in the execution and regulation of molecular processes. Yet little is known about the organization of mitochondrial proteins in such complexes.
Precision in the analysis of dynamic protein machines
The research groups of Thomas Becker and Fabian den Brave at the UKB, together with the research groups of Bernd Fakler, Uwe Schulte and Nikolaus Pfanner at the University of Freiburg, have created a high-resolution image of the organization of proteins in protein complexes, known as MitCOM. This involved a specific method known as complexome profiling to record the fingerprints of individual proteins at an unprecedented resolution. MitCOM reveals the organization into protein complexes of more than 90% of the mitochondrial proteins from baker’s yeast. This allows to identify new protein–protein interactions and protein complexes — an important information for further studies.
Quality control in the mitochondrial entry gate TOM as an example
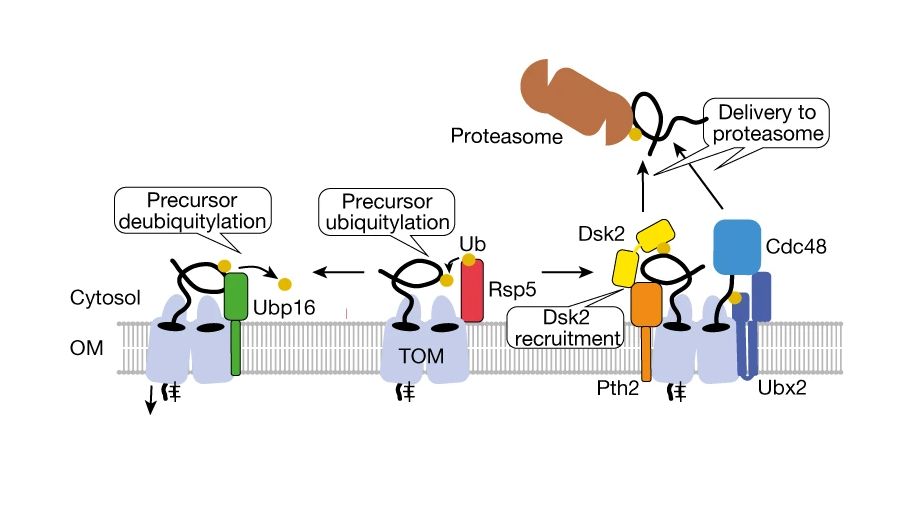
Researchers at UKB in cooperation with Collaborative Research Center 1218 (“Regulation of cellular function by mitochondria”) have shown how this dataset can be used to elucidate new processes. Mitochondria import 99% of their proteins from the liquid portion of the cell, known as cytosol. In this process, a protein machinery called the TOM complex enables the uptake of these proteins through the membrane into the mitochondria. However, it is largely unclear how proteins are removed from the TOM complex when they get stuck during the transport process. To elucidate this, the team led by Becker and den Brave used information from the MitCOM dataset. It was shown that non-imported proteins are specifically tagged for cellular degradation. Research by the Ph.D. student Arushi Gupta further revealed a pathway by which these tagged proteins are subsequently targeted for degradation. Understanding these processes is important because defects in protein import can lead to cellular damage and neurological diseases.
“The example from our study demonstrates the great potential of the MitCOM dataset to elucidate new mechanisms and pathways. Thus, this map of proteins represents an important source of information for further studies that will help us to understand the functions and origin of the cell’s powerhouse,” said Becker, director of the Institute of Biochemistry and Molecular Biology at UKB.
This article was first published by University Hospital Bonn. Read the original.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
CRISPR epigenome editor offers potential gene therapies
Scientists from the University of California, Berkeley, created a system to modify the methylation patterns in neurons. They presented their findings at ASBMB 2025.

Finding a symphony among complex molecules
MOSAIC scholar Stanna Dorn uses total synthesis to recreate rare bacterial natural products with potential therapeutic applications.

E-cigarettes drive irreversible lung damage via free radicals
E-cigarettes are often thought to be safer because they lack many of the carcinogens found in tobacco cigarettes. However, scientists recently found that exposure to e-cigarette vapor can cause severe, irreversible lung damage.

Using DNA barcodes to capture local biodiversity
Undergraduate at the University of California, Santa Barbara, leads citizen science initiative to engage the public in DNA barcoding to catalog local biodiversity, fostering community involvement in science.

Targeting Toxoplasma parasites and their protein accomplices
Researchers identify that a Toxoplasma gondii enzyme drives parasite's survival. Read more about this recent study from the Journal of Lipid Research.

Scavenger protein receptor aids the transport of lipoproteins
Scientists elucidated how two major splice variants of scavenger receptors affect cellular localization in endothelial cells. Read more about this recent study from the Journal of Lipid Research.

