From the journals: November 2018
We offer a selection of recent papers on a variety of topics from the Journal of Biological Chemistry, the Journal of Lipid Research and Molecular & Cellular Proteomics.
Proteomics of fish evolution
 This 1994 postage stamp from the Faroe Islands in the northern Atlantic Ocean shows an illustration of three-spined sticklebacks. Courtesy of Astrid Andreasen/
This 1994 postage stamp from the Faroe Islands in the northern Atlantic Ocean shows an illustration of three-spined sticklebacks. Courtesy of Astrid Andreasen/
Wikimedia Commons Ten thousand years ago, the ancestor of the three-spined stickleback lived only in the ocean. Today, its descendants have adapted to colonize many fresh, brackish and saltwater habitats, an evolutionary phenomenon known as adaptive radiation. Because of its recent, rapid diversification, the stickleback is used widely as a model organism in evolution and ecology research.
Researchers at the University of California, Davis, and Mexico’s Centro de Investigación en Alimentación y Desarrollo are reporting a new approach to understanding stickleback evolution. The researchers developed an assay using data-independent acquisition mass spectrometry to quantitate the most abundant proteins in gill tissue. They used the approach to compare four stickleback populations from environments ranging from Baja California to Anchorage, Alaska, that vary in water salinity, temperature and parasite exposure. Their research was published in Molecular & Cellular Proteomics.
Even in a small number of fish from each location, Johnathon Li and colleagues were able to identify proteomic signatures that appear to reflect adaptations to living in each location. The researchers anticipate that the proteomic assay will be useful for complex ecological modeling in future studies.
— Laurel Oldach
The complex life cycle of bones
Lateral meningocele syndrome, also known as Lehman syndrome, is a rare genetic disorder affecting the musculoskeletal system. The disorder is associated with gain-of-function mutations in the gene for the transmembrane receptor Notch3. Ernesto Canalis and colleagues at UConn Health examined the effects of these mutations on the mouse skeleton to understand the role of Notch3 in bone formation. They found that the mutant Notch3 triggered increased osteoblast proliferation, which counterintuitively resulted in reduced bone volume because of increased bone resorption. The findings were published in the Journal of Biological Chemistry.
Structure determines bacterial virulence
Campylobacter concisus and Campylobacter jejuni are both gram-negative bacteria that occupy the same ecological niche. While C. jejuni has been studied extensively in the pathogenesis of gastrointestinal diseases, there is very little information regarding the pathogenicity of C. concisus in such disorders. In a recent study published in the Journal of Lipid Research, Katja Brunner and colleagues at University College London investigated the structural differences of lipooligosaccharide, a common virulence factor known as LOS, between the two bacterial species. Through extensive mass spectrometry studies, the authors showed that the backbone of LOS in C. concisus is less phosphorylated and lacks sialylation, which hampers the interaction of the bacteria with a transmembrane protein that causes adherence, invasion and production of pro-inflammatory mediators. This study provides insight to differences in the LOS structure of these two organisms and how that correlates with their virulence.
Arctic insights into rubisco
The CO2-fixing enzyme rubisco is the most abundant enzyme on earth, but its catalytic properties vary among species. Marine phytoplankton generally have some of the most efficient rubisco enzymes known. Among the phytoplankton, diatoms are major players in the marine carbon cycle, but their rubisco enzymes have not been characterized. Karin Valegård and colleagues at Uppsala University showed that rubisco enzymes from five arctic diatom species were heavily post-translationally modified, suggesting that the role of these modifications in rubisco function should be investigated further. The findings were published in the Journal of Biological Chemistry.
How a common drug causes liver failure
Acetaminophen is a common pain reliever found in every pharmacy. However, it is also the No. 1 cause of acute liver failure in the United States. In the liver, acetaminophen is converted into a new compound that covalently binds to proteins with thiol groups. These covalent binding events contribute to the toxicity of acetaminophen, but they do not account fully for its role in liver failure.
James Chun Yip Chan and colleagues at the National University of Singapore examined a post-translational modification known as glutathionylation to understand its role in acetaminophen toxicity. In a study published in the journal Molecular & Cellular Proteomics, they showed that acetaminophen induces protein glutathionylation, which leads to mitochondrial dysfunction, deficits in energy metabolism and other effects linked to acetaminophen toxicity. This research helps explain a potential marker of drug toxicity and may be used in the future to reduce cases of liver failure worldwide.
Understanding an anti-malarial target
Efforts to control malaria are thwarted by drug resistance, motivating the search for new anti-malarial drugs. Several anti-malarial clinical candidates act on the ATPase PfATP4 in malaria parasites. PfATP4 is difficult to express heterologously, so its function has been poorly understood, and efforts to design better molecules that target it have been limited. James E.O. Rosling and colleagues at Australian National University developed a membrane ATPase assay to examine PfATP4’s biochemical properties. In the Journal of Biological Chemistry, they reported that PfATP4 is a pH-sensitive sodium transporter and identified the range of its sensitivity to an anti-malarial molecule under physiologically relevant conditions.
Steps forward for an anti-addictive plant compound
 Branches and fruit of an iboga shrub in the Limbe Botanical Garden, Cameroon. The plant’s bioactive compound, ibogaine, long has been discussed as a potential anti-addictive substance.Courtesy of Marco Schmidt/
Branches and fruit of an iboga shrub in the Limbe Botanical Garden, Cameroon. The plant’s bioactive compound, ibogaine, long has been discussed as a potential anti-addictive substance.Courtesy of Marco Schmidt/
Wikimedia CommonsThe opioid addiction crisis has revealed the need for safe treatments for withdrawal symptoms. The plant world is a promising source of psychoactive molecules that could be investigated for this purpose, but research into these drug candidates often is hampered by regulatory and technical roadblocks.
One plant with potential as an opioid addiction treatment is iboga (Tabernathe iboga), a shrub from west central Africa. In its native range, its root bark is used in traditional medicine and in rituals by practitioners of the Bwiti religion. In Western countries, iboga (and in particular its bioactive compound ibogaine) has been discussed as a potential anti-addictive substance since the 1960s, when the researcher and advocate Howard Lotsof used it to treat his own heroin addiction. Although clinical data are lacking and ibogaine remains unapproved for medical use, demand for the substance is sufficient that the iboga plant reportedly has become overharvested.
Hoping to help stabilize the ibogaine supply chain and facilitate research into ibogaine and related compounds, researchers at the John Innes Centre led by Scott C. Farrow are studying ibogaine biosynthesis.
The researchers produced the first transcriptome of the iboga plant. They then identified candidate genes that could be involved in ibogaine production by inferring comparisons to synthesis pathways of similar compounds in other plants. Ibogaine is a monoterpenoid indole alkaloid; alkaloids with similar structures (called iboga-type alkaloids) have been studied for their anti-cancer properties. By heterologously expressing candidate enzymes in yeast and E. coli with putative ibogaine precursors as substrates, the researchers showed that the two final enzymes in the pathway were a cytochrome P450 and an O-methyltransferase. They published their study in the Journal of Biological Chemistry.
The cytochrome P450, which the researchers named ibogamine-10-hydroxylase, is the first enzyme known to hydroxylate the iboga-type alkaloid scaffold, making it potentially a useful tool for developing new ibogaine derivatives. These findings, along with the publicly available ibogaine transcriptome, should enable further discovery of biosynthetic enzymes, leading to synthetic production of ibogaine and other bioactive molecules.
— Sasha Mushegian
Vitamin E formulations in low blood cholesterol
Chylomicron retention disease, known as CMRD, and abetalipoproteinemia, or ABL, are rare genetic disorders that cause impairment in absorption of fats and certain vitamins, resulting in hypocholesterolemia. One of the hallmarks of this group of diseases is severe vitamin E deficiency. Charlotte Cuerq and colleagues at Lyon Sud Hospital investigated the effect of two formulations of vitamin E (Tocofersolan and alpha-tocopherol acetate) on patients with CMRD and ABL in comparison to healthy volunteers.
In a paper published in the Journal of Lipid Research, they showed that in CMRD patients, Tocofersolan is better absorbed than the alternative formulation, as measured by a higher bioavailability after a single dose administration. To study how these two formulations change the absorption and storage of vitamin E, the authors administered the molecules for four months and found that vitamin E levels are higher in red blood cells, adipose tissue and plasma of patients with CMRD irrespective of the formulation. These observations are consistent with the less severe clinical phenotype in patients with CMRD than in patients with ABL.
Transcription factor allows parasite hijacking
Leishmania are intracellular parasites that deactivate host macrophages to evade the immune system. Previous evidence suggested that the parasite manipulated host miRNA as part of this process. In a paper in the Journal of Biological Chemistry, Lucie Colineau and colleagues at the University of British Columbia confirmed that Leishmania infection uniformly downregulated host cell miRNA expression. They also showed that this effect depended on expression of the host transcription factor c-Myc. Silencing c-Myc restored miRNA expression and reduced Leishmania’s intracellular survival.
Identifying bias in gene expression profiles
The compact nucleus of a cell houses all the information needed to make thousands of proteins. This information is found within genes that are organized carefully into clusters. Clustered genes often are transcribed into mRNA at the same time even though they may not be functionally related. However, research in cell lines has shown that cells can regulate expression of these unrelated genes at the protein level.
Piotr Grabowski and colleagues at the Institute of Biotechnology in Berlin examined factors that may contribute to the coexpression of functionally unrelated genes. In a study published in the journal Molecular & Cellular Proteomics, they describe how they used mouse tissues to show that epigenetics play an important role in regulating the coexpression of certain genes, but coexpression was buffered at the protein level. This research suggests that monitoring protein coexpression may be more useful than studying mRNA coexpression data for understanding functional genomics.
Solving the mystery of cholesterol deposits in the retina
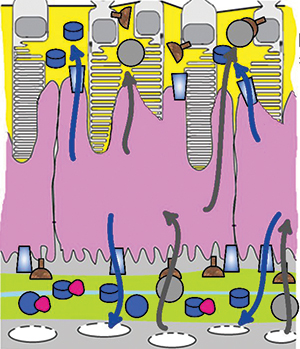 Cells in the retinal pigment epithelium (purple), which forms a barrier between photoreceptor cells (top/yellow) and capillaries (yellow-green), take up cholesterol from the bloodstream and export it in high-density lipoproteins (blue arrows) or in low-density lipoproteins (gray arrows). Raising the cholesterol available in the bloodstream increases the export of both types of lipoprotein toward the photoreceptors.Courtesy of Nicholas Lyssenko et al/
Cells in the retinal pigment epithelium (purple), which forms a barrier between photoreceptor cells (top/yellow) and capillaries (yellow-green), take up cholesterol from the bloodstream and export it in high-density lipoproteins (blue arrows) or in low-density lipoproteins (gray arrows). Raising the cholesterol available in the bloodstream increases the export of both types of lipoprotein toward the photoreceptors.Courtesy of Nicholas Lyssenko et al/
University of PennsylvaniaCholesterol deposits in the retina are linked to the onset of age-related macular degeneration, but it isn’t yet clear where the deposited cholesterol comes from. Some pathology studies suggest that it is secreted by the retinal pigment epithelium, or RPE, in particles similar to low-density lipoprotein, or LDL. But human genetic studies show a link between macular degeneration and genes involved in making high-density lipoprotein, or HDL. In a recent paper in the Journal of Lipid Research, Nicholas Lyssenko and colleagues at the University of Pennsylvania investigated HDL and LDL-like particle synthesis in the RPE.
The RPE forms a barrier between rod and cone photoreceptors and the capillaries that supply them with nutrients. Many epithelia actively control the nutrients that cross from the bloodstream into nearby tissues; the RPE does the same for the retina.
Researchers used a polarized tissue culture model of the RPE to trace isotope-labeled cholesterol. They observed that RPE cells were able to generate nascent HDL particles, but when ambient cholesterol was low, HDL synthesis was minimal, and cholesterol release occurred mainly through diffusion. When more cholesterol was available in the media, mimicking a person with elevated cholesterol levels, HDL synthesis was more robust, and RPE cells made more HDL at the photoreceptor side than at the capillary side. The authors also found that RPE cells secreted LDL-like particles weakly and mostly toward the photoreceptor side.
The research suggests that cholesterol deposited in the retina in age-related macular degeneration comes primarily from HDL secreted by the RPE.
— Laurel Oldach
A key enzyme in pseudoephedrine synthesis
Ephedrine and pseudoephedrine are pharmacologically important molecules produced by plants. The biosynthetic pathways in plants that produce these molecules are incompletely characterized, and commercial production of these molecules uses chemical rather than biological synthesis. In a study published in the Journal of Biological Chemistry, Jeremy S. Morris and colleagues at the University of Calgary identified the N-methyltransferase responsible for the final step in the synthesis of these molecules in Ephedra sinica. They then showed that this enzyme, when heterologously expressed in E. coli, was able to produce (pseudo)ephedrine, potentially paving the way for commercial production by engineered micro-organisms.
New opportunities to treat cholera
Cholera infection causes deadly dehydrating diarrhea, but the molecular details of how it does so are incompletely known. Diarrhea occurs when the chloride transporter cystic fibrosis transmembrane conductance regulator, or CFTR, is activated by cyclic adenosine monophosphate, or cAMP, in intestinal cells, but which biochemical pathway cAMP comes from in this context was unclear. Andrew Thomas and colleagues at Cincinnati Children’s Hospital Medical Center showed that the cAMP-generating adenylate cyclase AC6 is associated with CFTR in epithelial cells. When challenged with cholera toxin, cells in which AC6 was knocked out did not secrete fluid, suggesting that the AC6-CFTR complex could be a target for anti-diarrheal drugs. The findings were published in the Journal of Biological Chemistry.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Using DNA barcodes to capture local biodiversity
Undergraduate at the University of California, Santa Barbara, leads citizen science initiative to engage the public in DNA barcoding to catalog local biodiversity, fostering community involvement in science.

Targeting Toxoplasma parasites and their protein accomplices
Researchers identify that a Toxoplasma gondii enzyme drives parasite's survival. Read more about this recent study from the Journal of Lipid Research.

Scavenger protein receptor aids the transport of lipoproteins
Scientists elucidated how two major splice variants of scavenger receptors affect cellular localization in endothelial cells. Read more about this recent study from the Journal of Lipid Research.

Fat cells are a culprit in osteoporosis
Scientists reveal that lipid transfer from bone marrow adipocytes to osteoblasts impairs bone formation by downregulating osteogenic proteins and inducing ferroptosis. Read more about this recent study from the Journal of Lipid Research.

Unraveling oncogenesis: What makes cancer tick?
Learn about the ASBMB 2025 symposium on oncogenic hubs: chromatin regulatory and transcriptional complexes in cancer.

Exploring lipid metabolism: A journey through time and innovation
Recent lipid metabolism research has unveiled critical insights into lipid–protein interactions, offering potential therapeutic targets for metabolic and neurodegenerative diseases. Check out the latest in lipid science at the ASBMB annual meeting.


.jpg?lang=en-US&width=300&height=300&ext=.jpg)

