Identifying a new lipid metabolism gene
People with familial hypercholesterolemia, or FH, have very high levels of low-density lipoprotein cholesterol circulating in their blood due to aberrant LDL uptake by cells. With LDL levels elevated for prolonged periods, these patients are at increased risk for atherosclerotic cardiovascular disease.
“Mutations in several genes have been identified as contributors of FH,” Diego Lucero, a research fellow at the National Institutes of Health, explained. “However, a genetic link is still unidentified in about 20% to 40% of FH patients. This makes diagnosis and drug therapy design more challenging.”
Working in Alan T. Remaley’s lab, which focuses on understanding lipid metabolism and developing therapies to treat cardiovascular diseases, Lucero became interested in identifying other genes that contribute to aberrant LDL metabolism.
Through genomewide CRISPR–Cas9 knockout screening, Lucero used 76,441 sgRNAs to knock out 19,114 genes in Cas9-expressing HepG2 liver cells. sgRNA-transduced cells then were incubated with fluorescently labeled LDL and sorted for LDL uptake through flow cytometry. He collected cells with 5% or lower LDL uptake and deep sequenced them to determine sgRNA representation.
“If a gene influenced LDL uptake, its sgRNAs would feature among the most enriched in the deep sequencing,” Lucero said.
By studying sgRNA enrichment in his cell populations, Lucero identified 15 genes that influenced LDL uptake. He then generated HepG2 cell lines with these 15 candidate genes removed, and he remeasured LDL uptake in these cells.
“As expected, knockout of the LDLR gene showed the most robust reduction (about 80%) in cellular LDL uptake,” said Lucero. “We also observed consistent reductions in LDL uptake in three other novel genes.”
One of the three was transgelin.
Lucero worked with collaborators at the Mayo Clinic to validate the gene hits through the Global Lipids Genetics Consortium and lipid-related phenotypes available in UK Biobank. They found that differences in transgelin expression in human populations were associated strongly with elevated plasma lipids (triglycerides, total cholesterol and LDL cholesterol), making transgelin a target for further investigation. However, transgelin is an actin-binding protein that promotes motility in cells. What role does it play in lipid metabolism?
“In transgelin knockout cells, we found a universal 30% reduction in uptake of LDL, very low-density lipoprotein and transferrin,” Lucero said. “This led us to believe that transgelin affects something common between these cargos.”
When LDL binds to the LDL receptor, the latter is internalized, facilitating transport of LDL into the cell through clathrin-mediated endocytosis. And actin filament reorganization is a necessary step during clathrin-mediated endocytosis.
“Our microscopy experiments showed that transgelin plays a vital role during LDLR internalization, most likely by binding to actin filaments during endocytosis,” Lucero said. “This facilitates LDL uptake and consequently affects cellular cholesterol homeostasis.”
These findings recently were published in the Journal of Lipid Research. Lucero plans to continue this project using mice that are genetically modified to lack transgelin.
“We are also studying other proteins besides transgelin that might be involved in the uptake of LDL,” he said. “While this study focused on genes that reduce LDL uptake, we have also identified those that increase LDL uptake. This is an exciting direction because these might be therapeutic targets that could reduce cholesterol in blood.”
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.
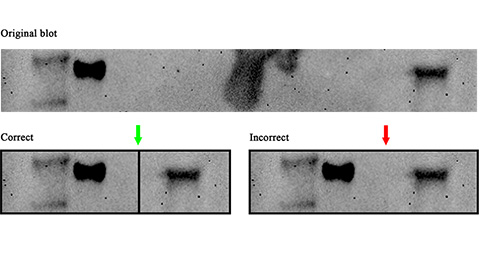
Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.
Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.
Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.
Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.