MCP: Charting the mitochondrial interactome
As the powerhouses of the cell, mitochondria host various supramolecular protein complexes. Delineating the structural basis of these protein complexes is essential to improve our understanding of how mitochondria function and generate energy. In a study published in Molecular & Cellular Proteomics, Albert Heck and a team of investigators at Utrecht University in the Netherlands, in collaboration with the National Institutes of Health, aimed to discover the organization and interactions of proteins in the mitochondria of mouse hearts.
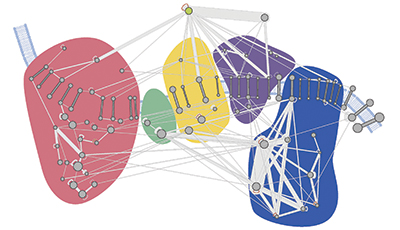 A molecular map of the oxidative phosphorylation supercomplexes. Courtesy of Philip Lössl/Utrecht University
A molecular map of the oxidative phosphorylation supercomplexes. Courtesy of Philip Lössl/Utrecht University
“We were most curious about the organization of protein molecules within mitochondria, because proteins are the molecular building blocks that make the mitochondrial energy factory work,” Heck said. “It was already known which proteins are involved in energy generation, but it is still not fully understood how these building blocks come together within intact mitochondria.”
To chart the organization of proteins within mitochondria, the researchers used a kind of molecular glue, or cross-linker, small enough to enter intact mitochondria and form stable links between any proteins within close proximity of one another. The mitochondria then were broken apart, and the proteins were digested and run on a mass spectrometer to identify especially the cross-linked peptides.
The researchers cataloged the largest set of mitochondrial protein interactions thus far, with 3,322 unique cross-links. This unprecedented depth was achieved using optimized mass spectrometry fragmentation schemes and data analysis strategies. “In contrast to earlier work based on similar strategies, our approach is much more sensitive, allowing us to present a more complete molecular interaction map of all proteins within mitochondria,” Heck said.
This molecular interaction map, or interactome, revealed a dense and interconnected network of proteins. The researchers used the map to study the higher-order organization of proteins and the architecture of protein complexes in mitochondria. Among these are the oxidative phosphorylation supercomplexes, a series of five protein complexes cumulatively responsible for generating energy. In addition to confirming known interactions, the researchers found novel cross-links between individual complexes, leading them to suggest that all five complexes coexist in close proximity.
Going a step further to validate their map, the investigators soaked the mitochondria in a high-salt solution to disrupt the protein supercomplexes. They showed, using the same cross-linking technique, that these “dysfunctional” mitochondria displayed a very different protein interaction network. “These data show that protein organization and mitochondria function are two sides of the same coin,” said co-author Philip Lössl. “We believe that our protein maps will help us understand the organization principles that allow mitochondria to work as molecular powerhouses.”
Chemical cross-linking and mass spectrometry allowed the researchers to probe native architecture of protein assemblies in mitochondria that are still intact and functioning. Most traditional biochemical methods for studying protein–protein interactions involve solubilizing the membrane using a detergent, which can introduce artifacts. “In such studies, the forceful breaking of the mitochondria can have dramatic effects on the protein organization and important information may be lost,” Lössl said. With the extensive comparative analyses and structural validation performed in their study, the researchers believe that the supercomplex interactions detected in intact mitochondria should be considered genuine.
The researchers believe their approach can be used to compare mitochondrial organization in diseases related to mitochondrial dysfunction, such as Parkinson’s and autism spectrum disorders. “Our approach can be used to elucidate how the molecular landscape of mitochondria is reprogrammed during disease development,” Heck said, “ultimately providing targets for future therapies.”
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

E-cigarettes drive irreversible lung damage via free radicals
E-cigarettes are often thought to be safer because they lack many of the carcinogens found in tobacco cigarettes. However, scientists recently found that exposure to e-cigarette vapor can cause severe, irreversible lung damage.

Using DNA barcodes to capture local biodiversity
Undergraduate at the University of California, Santa Barbara, leads citizen science initiative to engage the public in DNA barcoding to catalog local biodiversity, fostering community involvement in science.

Targeting Toxoplasma parasites and their protein accomplices
Researchers identify that a Toxoplasma gondii enzyme drives parasite's survival. Read more about this recent study from the Journal of Lipid Research.

Scavenger protein receptor aids the transport of lipoproteins
Scientists elucidated how two major splice variants of scavenger receptors affect cellular localization in endothelial cells. Read more about this recent study from the Journal of Lipid Research.

Fat cells are a culprit in osteoporosis
Scientists reveal that lipid transfer from bone marrow adipocytes to osteoblasts impairs bone formation by downregulating osteogenic proteins and inducing ferroptosis. Read more about this recent study from the Journal of Lipid Research.

Unraveling oncogenesis: What makes cancer tick?
Learn about the ASBMB 2025 symposium on oncogenic hubs: chromatin regulatory and transcriptional complexes in cancer.

