From the journals: MCP
Beyond microscopy: Mass spec's role in biopsies. Proteomic changes in learning and memory. Mapping mitochondria using mass spec. Read about papers on these topics recently published in the journal Molecular & Cellular Proteomics.
Beyond microscopy: Mass spec's role in biopsies
Microscopy is a standard method to assess biopsied tissue and look for abnormalities. This approach focuses largely on immunohistochemistry, or IHC, to identify and label cell lineage markers and therapeutic targets to provide insights for cancer diagnosis and treatment options. IHC has revolutionized cancer diagnosis and treatment, but it has faced significant obstacles in terms of standardization and consistency. While progress is being made to enhance IHC, its limited multiplexing capability and dependence on monospecific reagents may make it unsuitable for certain applications.
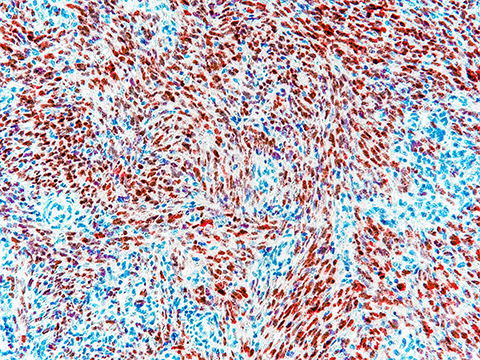
A recent perspective article in the journal Molecular & Cellular Proteomics by William Phipps and colleagues at the University of Washington and Fred Hutchinson Cancer Research Center discusses the challenges of using IHC for cancer diagnostics. It suggests that liquid chromatography–tandem mass spectrometry, or LC–MS/MS, might supplement biopsy testing methods. As laboratories use many different antibodies for IHC, antibody-based protein detection technologies may become less useful long-term; it may be hard to repeat results both within and between testing sites. Mass spectrometry could address conflicting results and supplement IHC.
The authors suggest that solutions using LC–MS/MS have several advantages, including increased specificity and sensitivity, decreased interference and turnaround time and the ability to multiplex. The authors stress that these methods would need to be appropriately calibrated, validated and consistently evaluated when used in the clinic.
Proteomic changes in learning and memory
Synaptic plasticity, the ability to change the strength of synapses, is important for learning and memory. An essential component of excitatory synapses is the postsynaptic density, or PSD, a dense network of hundreds of signaling molecules, neurotransmitter receptors and other proteins that control synaptic activity. Molecular and biological research has uncovered the identity and function of many of these proteins. However, researchers do not yet understand how the PSD changes during brain activities. Such insights could shed light on the neural networks that underlie memory and learning.
To address this gap in knowledge, Seok Heo and Taewook Kang of Johns Hopkins University and a group of researchers in Denmark examined the PSD proteome and phosphoproteome of mice after four different experiences, including inhibitory avoidance training, during which mice learn to associate the light or dark with an electrical shock. Their results were published in the journal Molecular & Cellular Proteomics.
The authors showed that the PSD levels of kinases, phosphatases and their phosphorylation status changed upon inhibitory avoidance training. Furthermore, they found that Ppm1h, a phosphatase that inhibits neuron elongation, regulates synaptic plasticity in cell culture and animals. Future research will focus on further defining the role of Ppm1h to determine if it is involved in disorders of memory or learning.
Mapping mitochondria using mass spec
Mitochondria are essential organelles packed full of proteins that provide a cell’s energy. Numerous disorders are associated with mitochondrial enzyme or protein complex malfunction, including mitochondrial encephalopathy and Leber hereditary optic neuropathy. Many of these proteins reside in separate subcompartments and physically interact with one another. This huge diversity of interactions makes it difficult to fully catalogue mitochondrial interactome.
Johannes F. Hevler and Albert J.R. Heck of the University of Utrecht recently published a perspective in the journal Molecular & Cellular Proteomics discussing the current state of crosslinking mass spectrometry, or XL-MS, and highlighting how it can be used to study the mitochondrial interactome Recent developments, such as cryogenic electron tomography, have further elucidated the mitochondria’s intricate membrane mazes. The authors assert that understanding these structures can be strengthened by XL-MS data, which uses distant restraints to reveal contact sites in protein–protein interactions. Other methods, such as affinity purification-mass spectrometry and identification by biotin ligation can further validate the interactions but are performed under less natural conditions.
The authors also discuss limitations of XL-MS, including low reaction efficiency, meaning that researchers cannot detect all protein interactions yet, especially that rare protein interactions may be missed. However, the authors argue that XL-MS is an ideal tool for discovering the mitochondrial interactome, despite these disadvantages.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Redefining excellence to drive equity and innovation
Donita Brady will receive the ASBMB Ruth Kirschstein Award for Maximizing Access in Science at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Mining microbes for rare earth solutions
Joseph Cotruvo, Jr., will receive the ASBMB Mildred Cohn Young Investigator Award at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Fueling healthier aging, connecting metabolism stress and time
Biochemist Melanie McReynolds investigates how metabolism and stress shape the aging process. Her research on NAD+, a molecule central to cellular energy, reveals how maintaining its balance could promote healthier, longer lives.

Mapping proteins, one side chain at a time
Roland Dunbrack Jr. will receive the ASBMB DeLano Award for Computational Biosciences at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Exploring the link between lipids and longevity
Meng Wang will present her work on metabolism and aging at the ASBMB Annual Meeting, March 7-10, just outside of Washington, D.C.

Defining a ‘crucial gatekeeper’ of lipid metabolism
George Carman receives the Herbert Tabor Research Award at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

