How bacteria inhibit gene expression
Bacteria adapt to environmental changes by regulating their metabolism. The best way to do this is to change the gene expression. Proteins called transcription factors take care of this job and help organisms respond to change.
The transcription factor CarD is conserved among several bacterial lineages and reacts with other proteins to prevent or initiate gene expression. Although researchers know the initiating mechanism of CarD, they do not yet understand how it inhibits gene expression. Understanding the complete mechanism of CarD activity may help scientists develop new approaches to combat bacteria.
Dennis Zhu and Christina Stallings, researchers at the Washington University School of Medicine, study the inhibiting mechanism in Mycobacterium tuberculosis, or Mtb. They recently showed which factors affect CarD’s activity in gene expression.
The RNA polymerase, or RNAP, enzyme binds to DNA to promote transcription. RNAP binds a promoter sequence to start transcription, and the RNAP–promoter interaction is called RNAP-promoter open complex, or RPo. Previous studies showed that CarD stabilizes RPo and initiates RNAP’s activity. However, using RNA sequencing, Zhu and Stallings showed that altering the activity of CarD can cause downregulation and upregulation of some genes.
According to Zhu, their findings published in the Journal of Biological Chemistry are broadly applicable for other bacterial transcription factors as well.
“I believe what makes our study unique is that we approached it with a constrained hypothesis that the outcome of CarD regulation on mycobacterial transcription is dependent on RPo stability,” Zhu said. “Other studies of RNAP-binding transcription factors have explored the natural promoter space of a bacterium and have performed similar promoter bashing to identify regulatory sequences.”
Unlike previous studies, Zhu and Stallings observed the effect of CarD under different promoters. They showed that it has an inhibitory effect on promoters that already have highly stable RPo. This is the first demonstration of CarD’s transcriptional repression activity, and they believe that this is how CarD inhibits some genes.
The researchers did all these experiments in the controlled conditions of a laboratory. However, a bacteria’s natural environment is different. “The main limitation of studies like ours is the gap in understanding between the next-generation sequencing data sets that we gather from bacteria and the mechanistic details from our in test tube assays,” Zhu said. “So many things are happening within the bacterium such as salt conditions, other proteins, chromosome architecture, translation, which we cannot fully capture in an in vitro transcription assay.”
The next step for the group is creating a bridge between findings in the lab and more natural settings. CarD is a stress-response protein. That is, when bacteria feel environmental stress such as lack of nutrition or DNA damage, CarD is downregulated or upregulated, respectively.
Zhu and colleagues got bulk RNA sequencing data sets by exponentially growing Mycobacterium tuberculosis cells in a nutrient-rich media. “It would be interesting to explore how certain stress conditions, particularly nutrient starvation and DNA damage, affect CarD’s ability to regulate,” he said.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
The data that did not fit
Brent Stockwell’s perseverance and work on the small molecule erastin led to the identification of ferroptosis, a regulated form of cell death with implications for cancer, neurodegeneration and infection.

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.
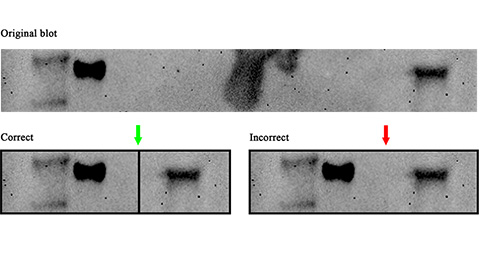
Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.
Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.
Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.
Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

