Mechanisms of a gut bacterium – and how to stop them
The human gastrointestinal tract is one of the most diverse and complex ecosystems, comprising trillions of microbes. Bile acids produced by the liver and secreted into the intestines not only aid in the digestion of food but also protect against pathogenic microorganisms in the gut. To survive the hostile environment of the GI tract, bacteria must develop strategies for resistance.
Vibrio parahaemolyticus, a Gram-negative bacterium that causes food-borne diseases can sense and use bile acids as a signal to activate harmful toxins that cause diarrhea. Understanding how pathogenic bacteria evade host defense mechanisms is critical in developing new treatments to prevent or cure gastrointestinal diseases.

Kim Orth, at the University of Texas Southwestern Medical Center, leads a team that researches virulence mechanisms used by V. parahaemolyticus to survive in the GI tract. In a recent study published in the Journal of Biological Chemistry, graduate student Angela Zhou and colleagues show how V. parahaemolyticus differentially senses bile acids to activate toxin production.
A major pathogenic factor of the organism is a type III secretion system called T3SS2. T3SS2 comprises bacterial structures that help inject effector proteins that enable the V. parahaemolyticus to invade the host and establish infection. “When Vibrio parahaemolyticus is ingested and enters the intestines, it senses bile acids and activates the T3SS2,” Orth said. “The T3SS2 secretion system injects toxins from the bacteria into cells lining the intestine, causing disease.”
Two membrane proteins in the bacteria, VtrA and Vtr-C, form a complex to control production of the T3SS2. The VtrA–VtrC responds to the host GI tract bile acids to induce VtrB, another membrane-bound protein that activates the T3SS2. Bile acids, such as taurodeoxycholate or TDC, trigger VtrB expression and T3SS2 activation, while others like chenodeoxycholate, or CDC, do not.
Using X-ray crystallography, isothermal titration calorimetry and green fluorescent protein reporter assays, Zhou and colleagues determined why TDC, but not CDC, activates the T3SS2 upon binding VtrA–VtrC. They found that the differences in sensing bile acids were due to the subtle differences in the molecular structures of CDC and TDC. They showed that two amino acids, histidine50 and serine123, were vital for determining whether TDC and not CDC activates the T3SS2.
“These bile acids act like a card reader for a locked door,” Orth said. “If a person has access to the room behind the door, inserting their card into the card reader will unlock the door, and if a person who does not have access inserts their card, the door will remain locked,” Orth added.
The team was excited to find that CDC also binds to VtrA–VtrC within the same pocket in the protein as TDC. However, a closer examination of the structure's atomic-level interactions showed that CDC and TDC bind to VtrA–VtrC differently.
“Our research reveals how enteric bacteria use environmental cues to cause gastrointestinal disease and provides clues on why certain people are more susceptible to disease than others,” Orth said. "Researchers recently found bile acids are modified by good bacteria that live in the intestines, which can protect against harmful bacteria by changing the makeup of the bile acid pool in the intestines.”
The researchers performed the X-ray crystallography and ITC experiments on only a part of the VtrA and VtrC proteins, the periplasmic domain. In the future, the team will focus on determining the molecular mechanism of differential activation of the T3SS2 by TDC and CDC using the entire VtrA and VtrC proteins.Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
The data that did not fit
Brent Stockwell’s perseverance and work on the small molecule erastin led to the identification of ferroptosis, a regulated form of cell death with implications for cancer, neurodegeneration and infection.

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.
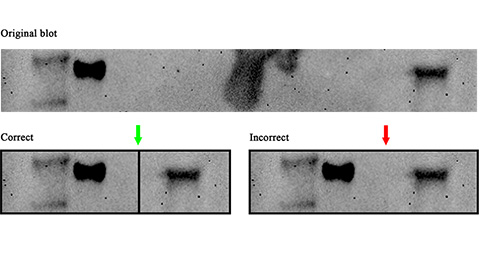
Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.
Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.
Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.
Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

