Nobel for ‘breakthrough in biochemistry’
The 2024 Nobel Prize in chemistry today was awarded to David Baker “for computational protein design” and to Demis Hassabis and John M. Jumper “for protein structure prediction.”



“One of the discoveries being recognized this year concerns the construction of spectacular proteins,” Heiner Linke, chair of the Nobel Committee for Chemistry 2024, said in a press release. “The other is about fulfilling a 50-year-old dream: predicting protein structures from their amino acid sequences. Both of these discoveries open up vast possibilities.”
Baker, a professor at the University of Washington, used traditional computer algorithms to pioneer a method to design proteins that do not exist in nature. His work opens avenues for designing pharmaceuticals, vaccines, nanomaterials and more.
Hassabis and Jumper, of Google DeepMind, developed multiple iterations of AlphaFold, a computer model that can predict protein structures from amino acid sequences. Using machine learning and artificial intelligence, they have predicted the structure of nearly all known proteins.
Collectively, the trio’s contributions will allow scientists to tackle 21st century challenges such as antibiotic resistance and plastic degradation.
F. Ulrich Hartl, a professor at the Max Planck Institute of Biochemistry and co-winner of the 2013 American Society of Biochemistry and Molecular Biology Tabor Award with Arthur Horwich, said the prize “could not be more highly deserved” and emphasized the far-reaching impacts of these discoveries.
“Together, these scientists have revolutionized structural biology. We can now predict with accuracy the shapes of existing proteins and make entirely new ones, with the potential to develop new functionalities for medical treatment and biotechnology. The methods and programs they have developed are now used in practically every lab working with proteins,” Hartl said.
Linke said the discoveries behind the 2024 prize led to a “breakthrough in biochemistry.”
“(M)aking the connection in both ways between amino acid sequence and protein structure, which then is related to protein function, has been defined,” Linke said in the Nobel press conference. “That was actually called a grand challenge in chemistry, and in particular in biochemistry, for decades.
Protein design
Baker earned his Ph.D. from the University of California, Berkeley, in the laboratory of Randy Schekman, who won the 2013 Nobel Prize in physiology or medicine for discovering vesicular trafficking.
“(Baker’s) energy, enthusiasm and brilliance transformed the work in my lab to allow a biochemical approach to advance our genetic analysis of the mechanism of protein secretion,” Schekman said. “Baker’s success is no accident. He has the rare gift of creativity and confidence to make magic happen in the lab.”
After opening his own lab, Baker used his knowledge of life’s building blocks to design a novel protein that doesn’t exist in nature, known as Top7, in 2003.
“As we got better and better at (designing proteins), the scope of applications became more and more exciting,” Baker said. “It's been this huge opening up of possibilities, because the proteins in nature do so many different things. They mediate all the processes in our body and in all living things — and the beginning that it might be possible to create a whole new world of proteins that could address a lot of the problems faced by humans in the 21st century.”
Baker used the Rosetta computer program, which he developed in 1999, to assemble structural fragments from unrelated protein structures with similar local sequences in the Protein Data Bank and simultaneously optimize sequence and structure.
Janet Smith, a professor of biophysics at the University of Michigan Life Sciences Institute and winner of the 2022 ASBMB Mildred Cohn Award in Biological Chemistry, offered her congratulations.
“David Baker has transformed the field of protein design through his creative design of entirely new proteins with real-world applications that he demonstrates by moving design projects out of the computer and into the lab,” Smith said.
In 2023, Baker attended the ASBMB annual meeting and presented his work on protein design using deep learning at a symposium about AI and machine learning in structural biology. One of the symposium chairs was Celia Schiffer, professor and chair of biochemistry and molecular biotechnology at the University of Massachusetts Chan Medical School and winner of the 2020/2021 ASBMB William C. Rose Award.
“Basically, David cracked the protein folding code, developing artificial intelligence computational techniques — and validated them with experimental structures — of how amino acid sequences can define a specific three-dimensional protein structure and ultimately function,” Schiffer said.
During the Nobel press conference, a reporter asked Baker about his favorite protein.
“I love all the proteins, so I don't want to pick a favorite, but I can tell you about one that we designed during the pandemic that protects against coronavirus,” Baker said.
During the COVID-19 pandemic, Baker’s group collaborated with scientists including Michael Jewett, a professor of bioengineering at Stanford University, to design “miniprotein inhibitors” to block the SARS-CoV-2 virus’ interaction with its receptor, angiotensin-converting enzyme 2. Their most potent inhibitor neutralized the virus with similar or greater potency than antibody treatments with Emergency Use Authorization status from the U.S. Food and Drug Administration.
“These innovations change the way people think about proteins and how they do protein science,” Jewett said. “Success in harnessing AI and computational predictions of protein structure is bringing new solutions to address some of society's greatest challenges in human and planet health.”
Baker and collaborators created a nasal spray containing that potent novel miniprotein inhibitor, and it is now in clinical trials for protecting against SARS-CoV-2.
“I've been very excited about the idea of a nasal spray of little designer proteins that would protect against all possible pandemic viruses,” he said.
Jewett added that Baker’s work has inspired many. “He is an inspirational leader, collaborator and mentor. He has built an enduring and supportive community and shown how single-minded dedication to a problem and just loving science can lead to incredibly transformative results.”
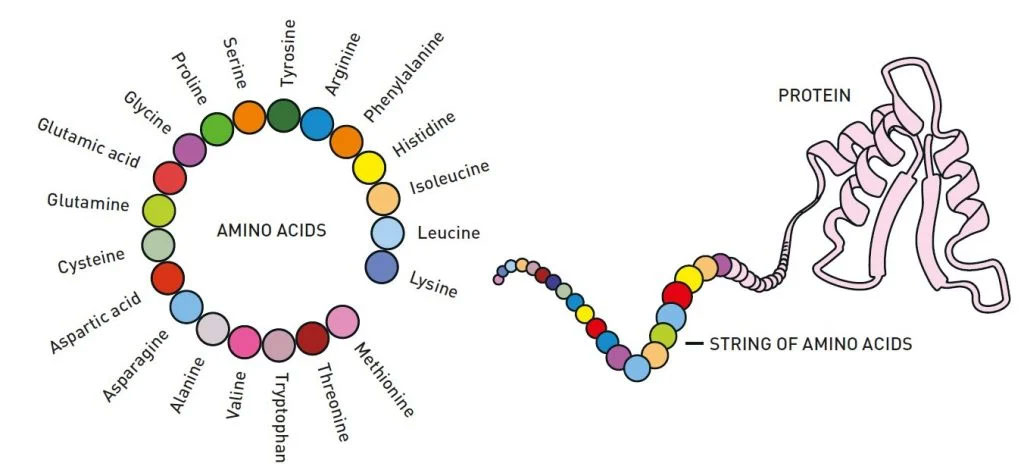
Protein folding
The second half of the chemistry prize went to Hassabis and Jumper at Google DeepMind, an artificial intelligence research laboratory.
Prior to 2016, scientists could predict protein structures with only about 40% accuracy.
“Ever since 1962, when Chris Anfinson showed the world that the 3D structure of a protein is somehow encoded in the amino acid sequence, ‘the protein-folding problem’ has been one of the great intellectual challenges of biology,” Smith said.
In 2019, Hassabis and Jumper released AlphaFold, a protein-structure prediction program, using convolutional neural networks and structures found in the Protein Data Bank. However, this program predicted structures with only 60% accuracy.
In 2020, they made a breakthrough. They designed a second iteration of AlphaFold using PDB and neural network machine learning, for which John Hopfield and Geoffrey Hinton on Tuesday won the 2024 Nobel Prize in physics. The program, known as AlphaFold2, predicted protein structures with an unprecedented 90% accuracy within a backbone accuracy of about 1Å.
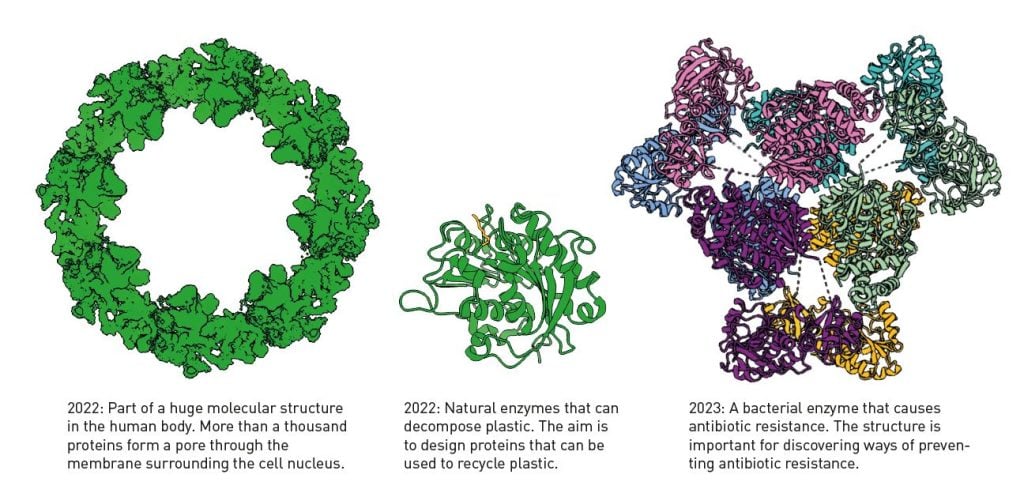
Hassabis and Jumper improved the algorithm behind AlphaFold2 and released AlphaFold3 in 2024. This tool allows researchers to predict protein complexation with DNA, RNA, ligands and ions. In addition, the tool can predict structures with posttranslational modifications.
Thanks to Hassabis and Jumper, free databases now hold over 200 million proteins from 100 million species and 48 complete proteomes, including humans and model organisms.
Because Hassabis and Jumper made the AlphaFold2 source code freely available, researchers such as Smith and Baker are using it to advance their own research.
“AlphaFold has transformed molecular biology,” Smith said. “The predicted structures are (mostly) reliable enough that biologists can use them as a basis for experiments.”
Baker described the power of artificial intelligence, or AI, as “tremendous” and said he is excited to see how this technology plays a role in the future of society.
“I'm really excited about all the ways in which protein design can now make the world a better place, in health, medicine and really, outside technology, sustainability.”
Karen Fleming, a professor of biophysics at Johns Hopkins University and president-elect of the Biophysical Society, agreed, saying: “These tools are game changers for science and medicine and probably many other applications we haven’t even thought of yet.”
These discoveries have also changed how educators teach biochemistry on college campuses.
“In our biochemistry lab course, students use computational methods with sequence and structure information to predict the function of protein structures that have not been annotated, then move into the wet lab to test their predictions,” Paul Craig, professor of biochemistry at the Rochester Institute of Technology, recipient of the 2018 ASBMB Award for Exemplary Contributions to Education and ASBMB fellow, said. “Until recently we were limited to protein structures generated by the Structural Genomics Initiative, around 4,000 structures in the PDB. With the advent of computed structure models, we now have an almost unlimited source of proteins with little or no annotation for our students to explore.”
Even though the programs and data generated by the 2024 laureates have changed the way scientists study proteins, Tanja Mittag, a member of the structural biology department at St. Jude Children’s Research Hospital, said there is more work to be done on protein design and structure prediction.
“Obviously these two approaches do not solve all protein biochemistry problems,” Mittag said. “But they bring us large steps forward. And importantly, they also show us what is possible with lots of high-quality data.”
Neil Kelleher, a professor of life sciences at Northwestern University, agreed.
“After the genome era, the recognition of computational protein design with a 2024 Nobel Prize shows just how far we have moved to understand and manipulate protein-level chemistry,” Kelleher said. “However, protein-level biology still has a major leap, and we don’t have some of the basics in place: for example, knowledge of the actual full-length protein molecules that occur in a multitude of protein forms, now called proteoforms. Moving forward, deep proteoform sequencing will be required to ‘domesticate’ the proteome and achieve precision medicine through a combination of long-read proteomics and multi-omics.”
The laureates will share a prize fund worth 11 million Swedish kronor, or approximately $1 million.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Targeting Toxoplasma parasites and their protein accomplices
Researchers identify that a Toxoplasma gondii enzyme drives parasite's survival. Read more about this recent study from the Journal of Lipid Research.

Scavenger protein receptor aids the transport of lipoproteins
Scientists elucidated how two major splice variants of scavenger receptors affect cellular localization in endothelial cells. Read more about this recent study from the Journal of Lipid Research.

Fat cells are a culprit in osteoporosis
Scientists reveal that lipid transfer from bone marrow adipocytes to osteoblasts impairs bone formation by downregulating osteogenic proteins and inducing ferroptosis. Read more about this recent study from the Journal of Lipid Research.

Unraveling oncogenesis: What makes cancer tick?
Learn about the ASBMB 2025 symposium on oncogenic hubs: chromatin regulatory and transcriptional complexes in cancer.

Exploring lipid metabolism: A journey through time and innovation
Recent lipid metabolism research has unveiled critical insights into lipid–protein interactions, offering potential therapeutic targets for metabolic and neurodegenerative diseases. Check out the latest in lipid science at the ASBMB annual meeting.
Melissa Moore to speak at ASBMB 2025
Richard Silverman and Melissa Moore are the featured speakers at the ASBMB annual meeting to be held April 12-15 in Chicago.

