Engineering cells to broadcast their behavior can help scientists study their inner workings
Waves are ubiquitous in nature and technology. Whether it’s the rise and fall of ocean tides or the swinging of a clock’s pendulum, the predictable rhythms of waves create a signal that is easy to track and distinguish from other types of signals.
Electronic devices use radio waves to send and receive data, like your laptop and Wi-Fi router or cellphone and cell tower. Similarly, scientists can use a different type of wave to transmit a different type of data: signals from the invisible processes and dynamics underlying how cells make decisions.
I am a synthetic biologist, and my research group developed a technology that sends a wave of engineered proteins traveling through a human cell to provide a window into the hidden activities that power cells when they’re healthy and harm cells when they go haywire.
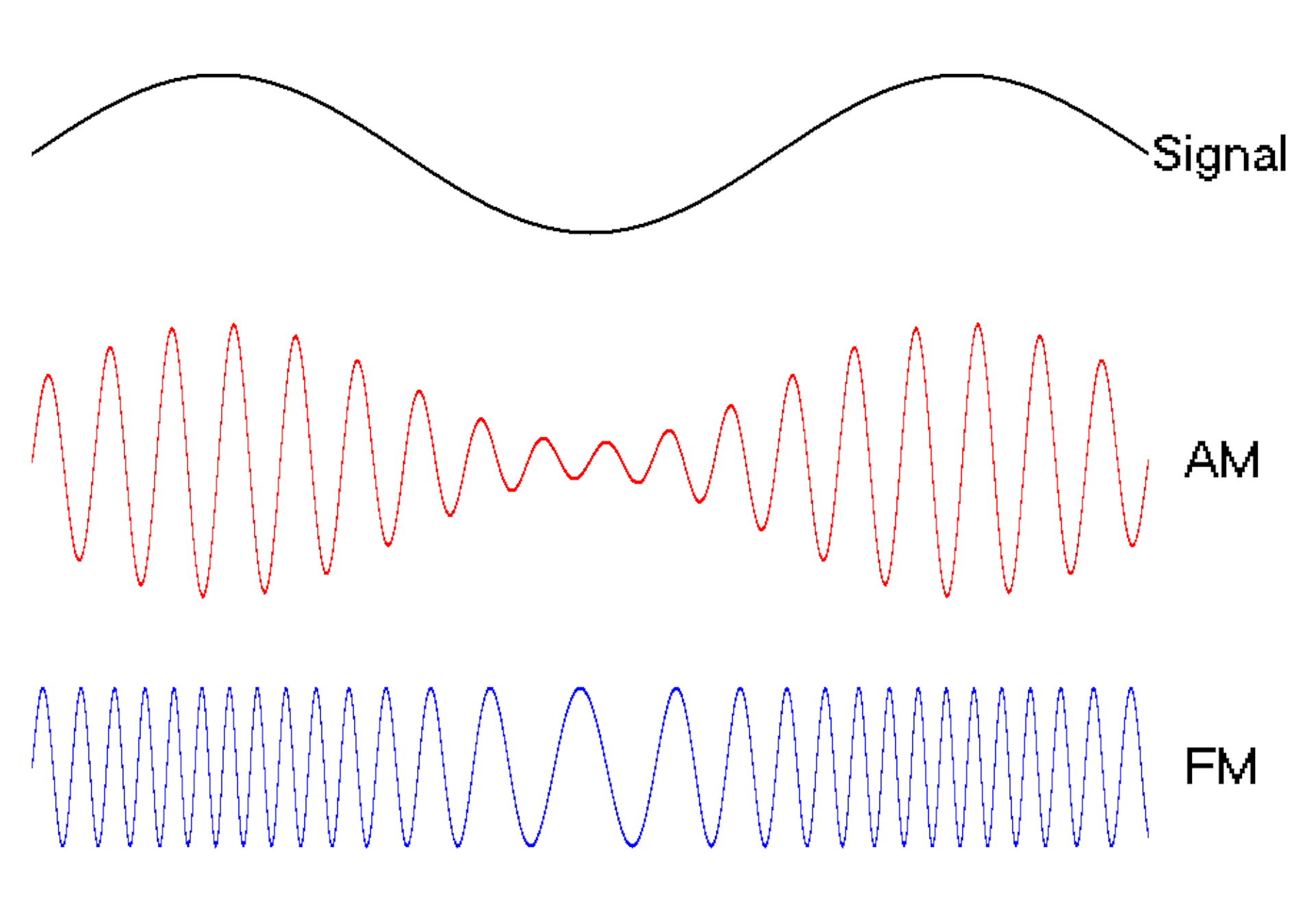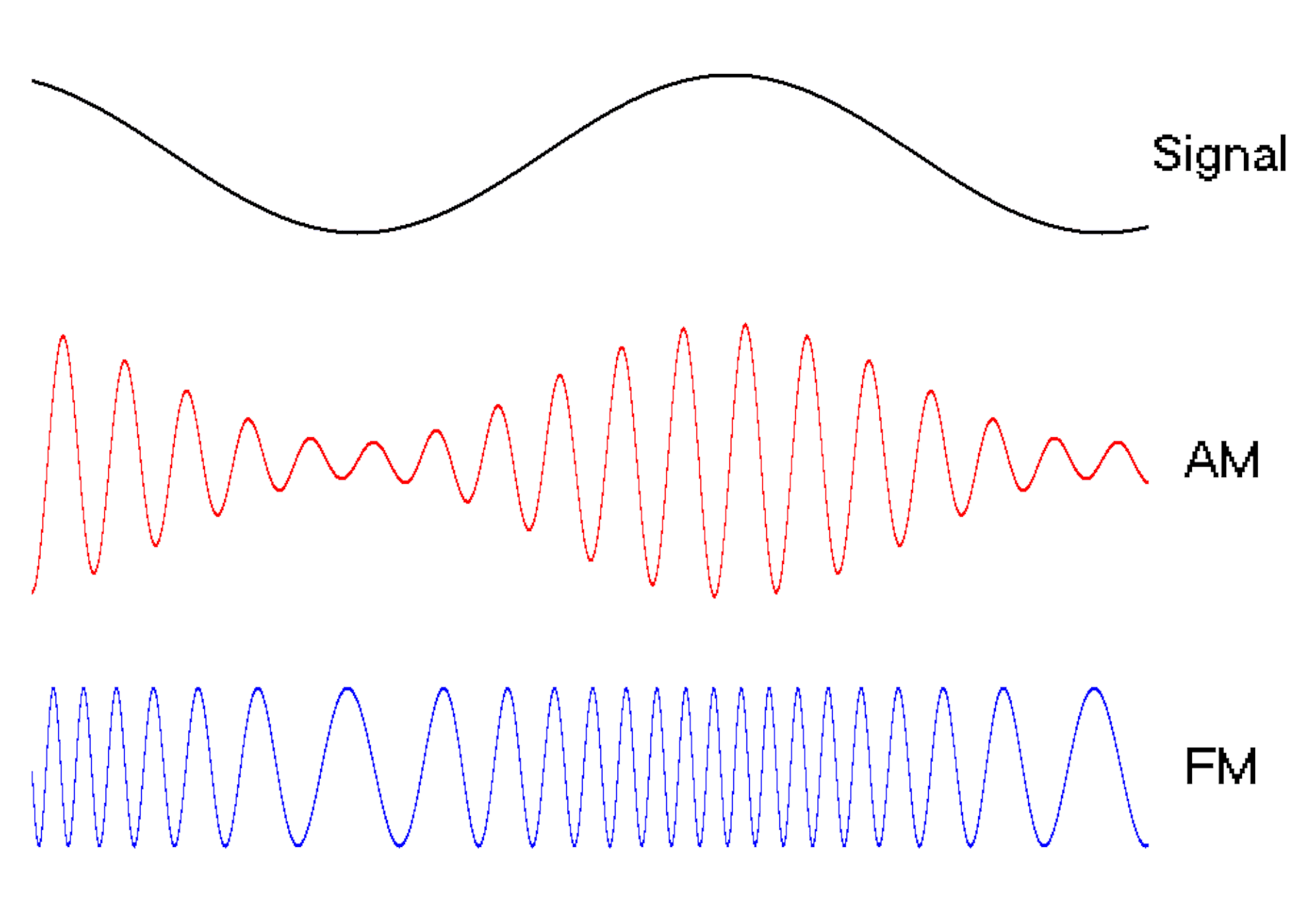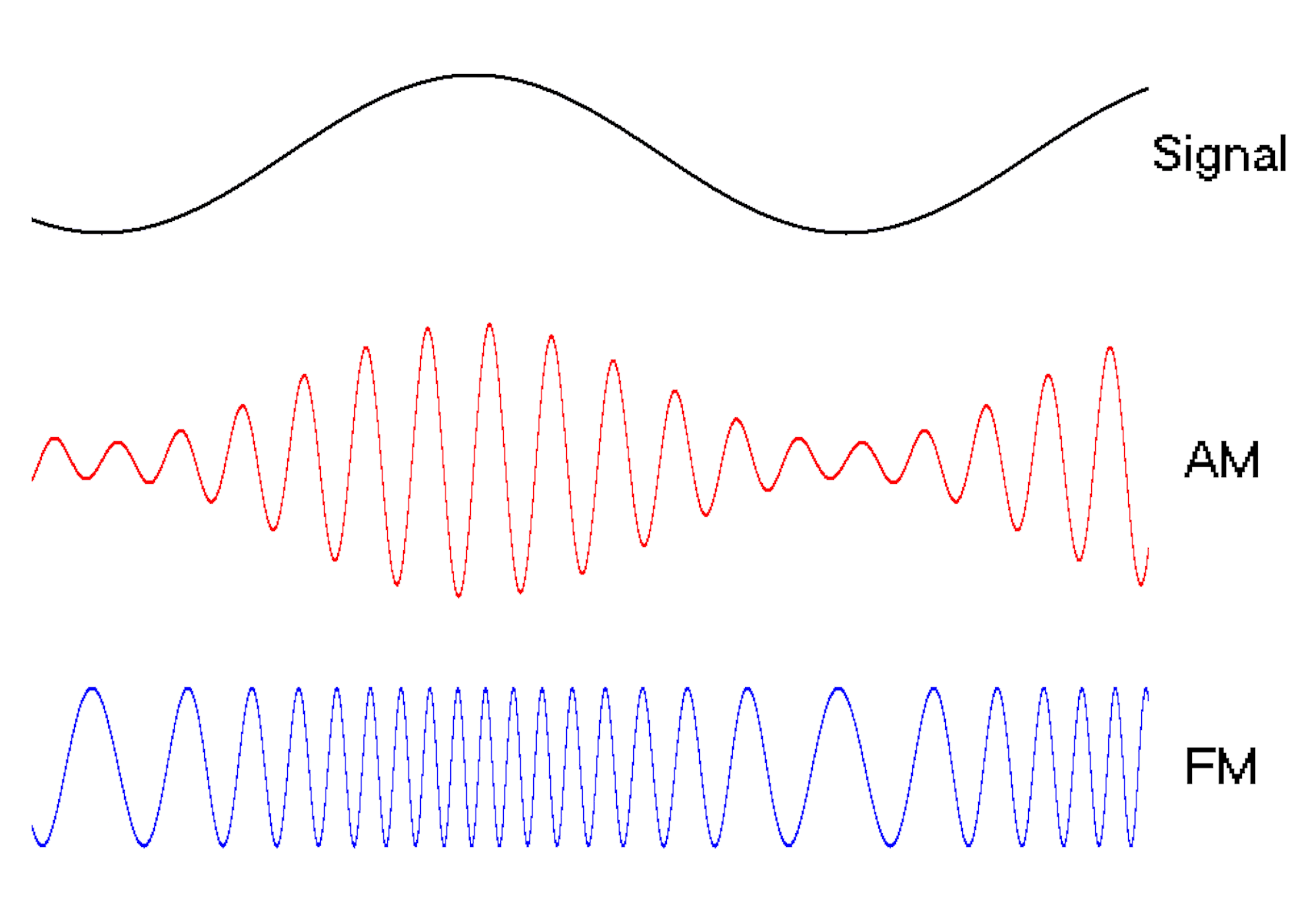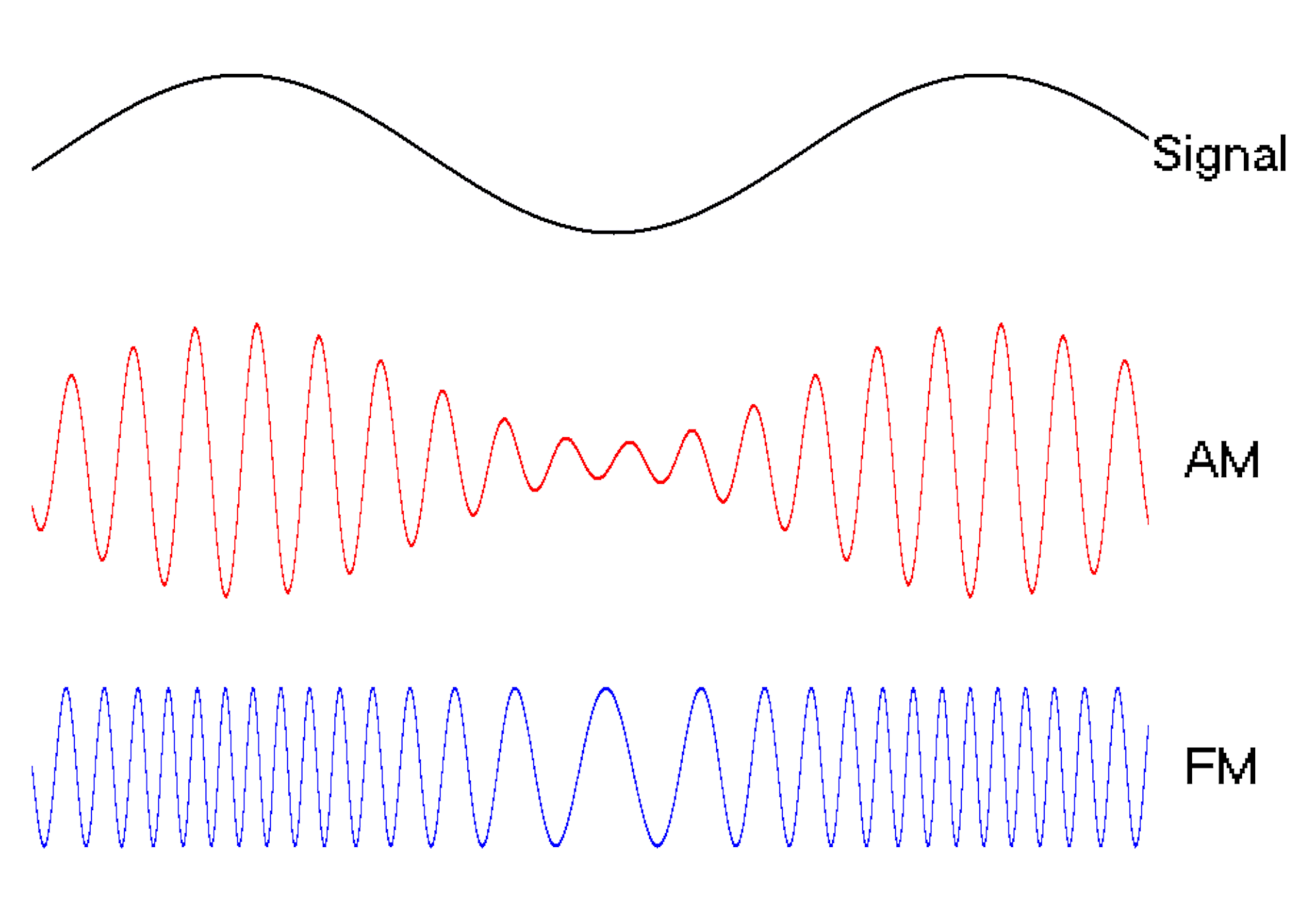
Waves are a powerful engineering tool
The oscillating behavior of waves is one reason they’re powerful patterns in engineering.
For example, controlled and predictable changes to wave oscillations can be used to encode data, such as voice or video information. In the case of radio, each station is assigned a unique electromagnetic wave that oscillates at its own frequency. These are the numbers you see on the radio dial.
Scientists can extend this strategy to living cells. My team used waves of proteins to turn a cell into a microscopic radio station, broadcasting data about its activity in real time to study its behavior.
Turning cells into radio stations
Studying the inside of cells requires a kind of wave that can specifically connect to and interact with the machinery and components of a cell.
While electronic devices are built from wires and transistors, cells are built from and controlled by a diverse collection of chemical building blocks called proteins. Proteins perform an array of functions within the cell, from extracting energy from sugar to deciding whether the cell should grow.
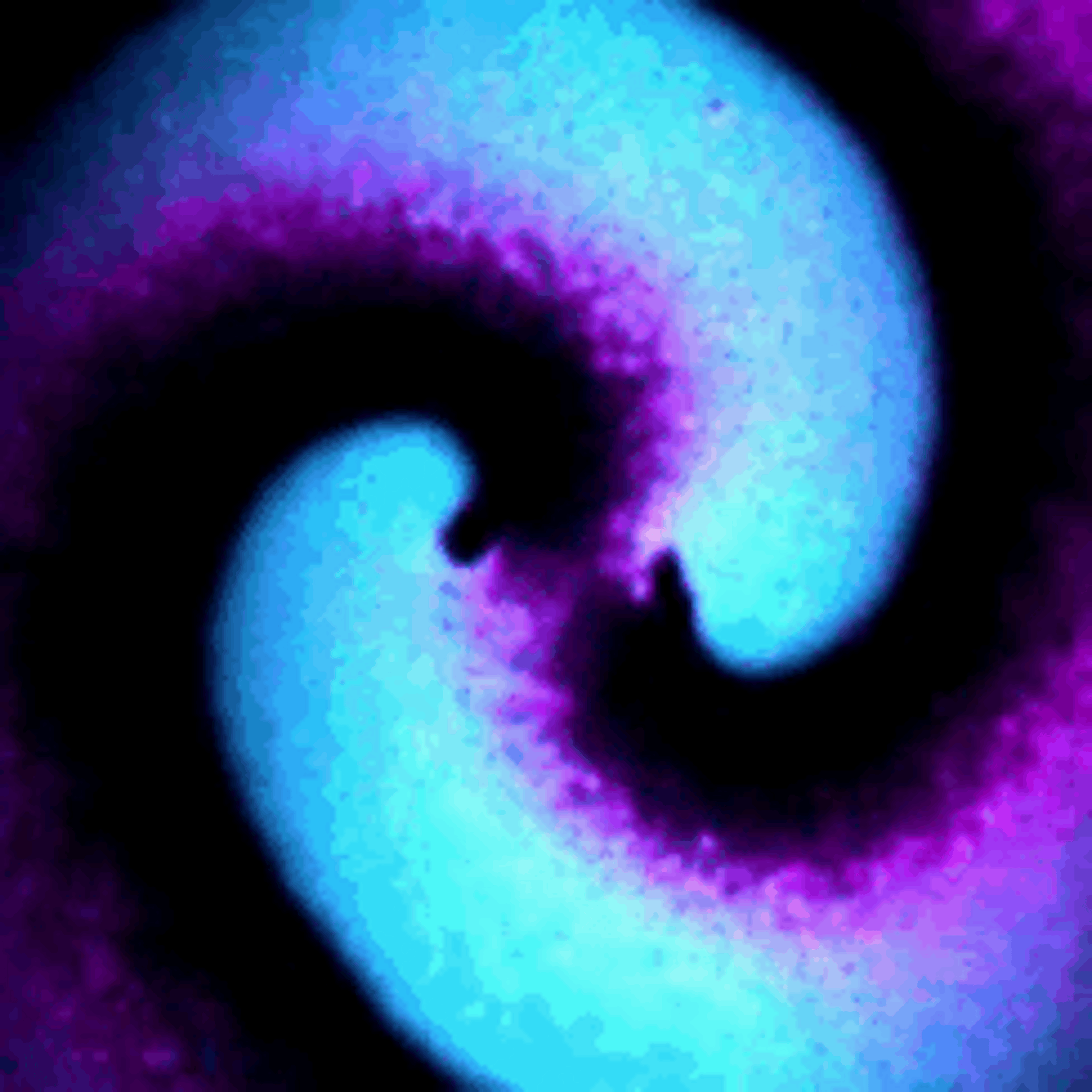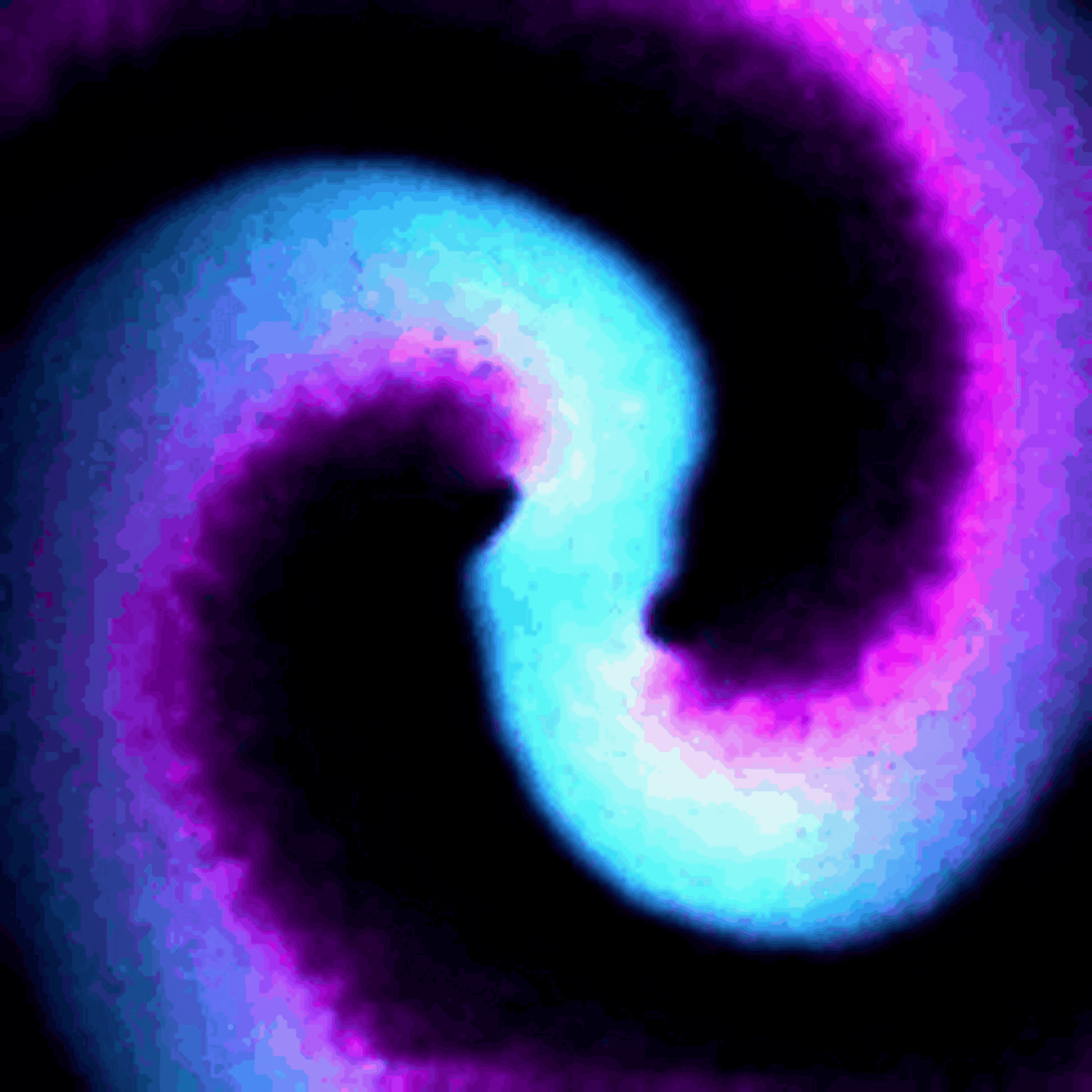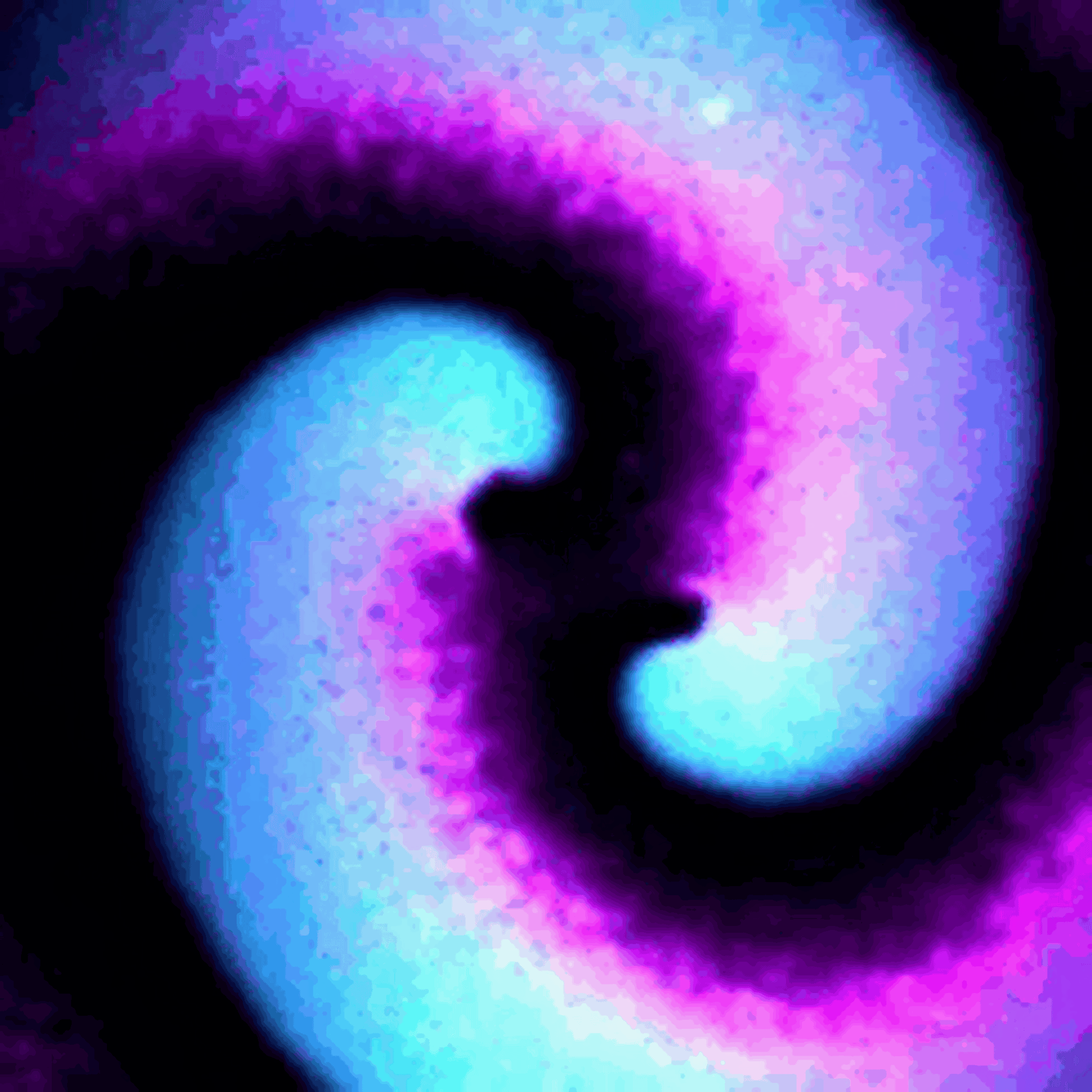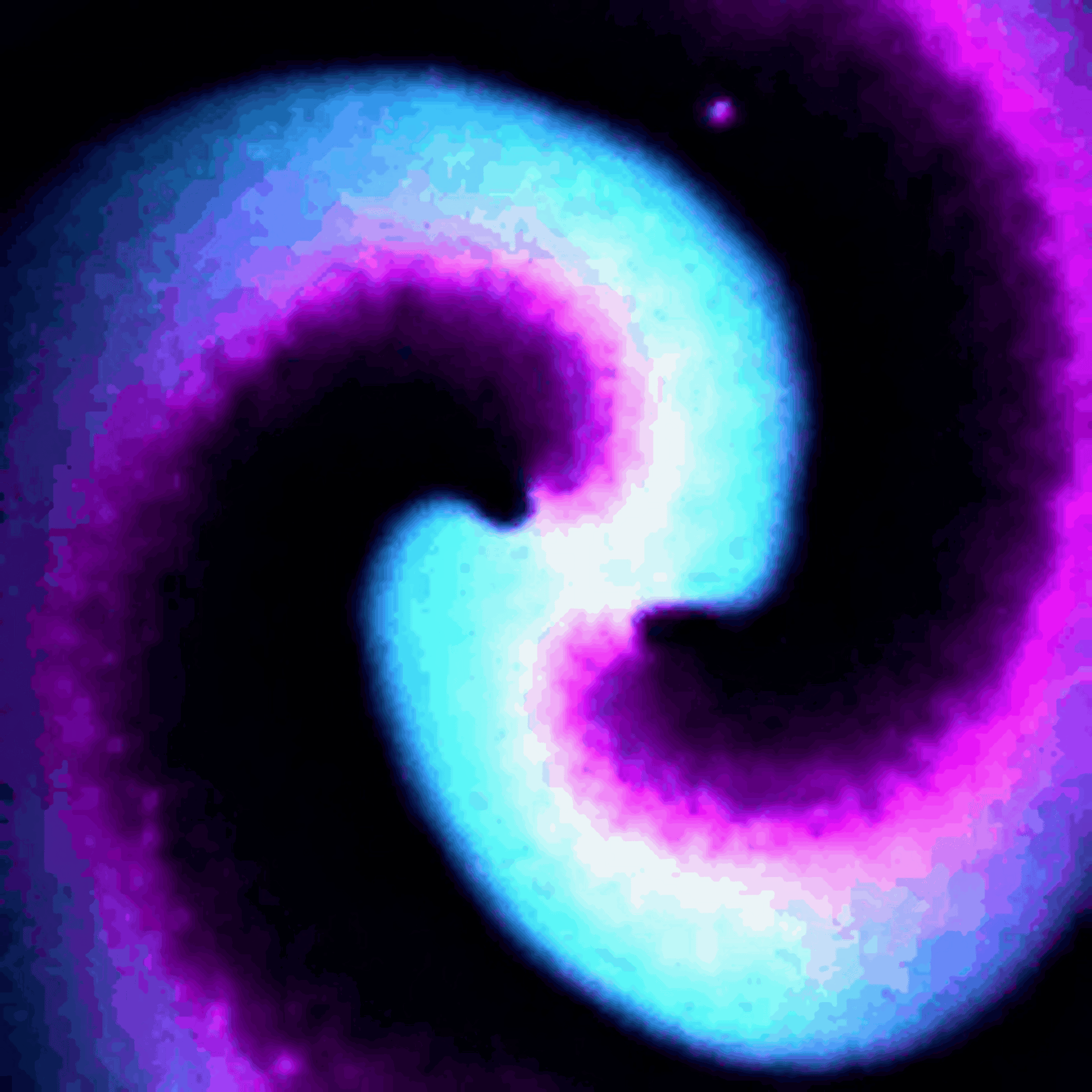
Protein waves are generally rare in nature, but some bacteria naturally generate waves of two proteins called MinD and MinE – typically referred to together as MinDE – to help them divide. My team discovered that putting MinDE into human cells causes the proteins to reorganize themselves into a stunning array of waves and patterns.
On their own, MinDE protein waves do not interact with other proteins in human cells. However, we found that MinDE could be readily engineered to react to the activity of specific human proteins responsible for making decisions about whether to grow, send signals to neighboring cells, move around and divide.
The protein dynamics driving these cellular functions are typically difficult to detect and study in living cells because the activity of proteins is generally invisible to even high-power microscopes. The disruption of these protein patterns is at the core of many cancers and developmental disorders.
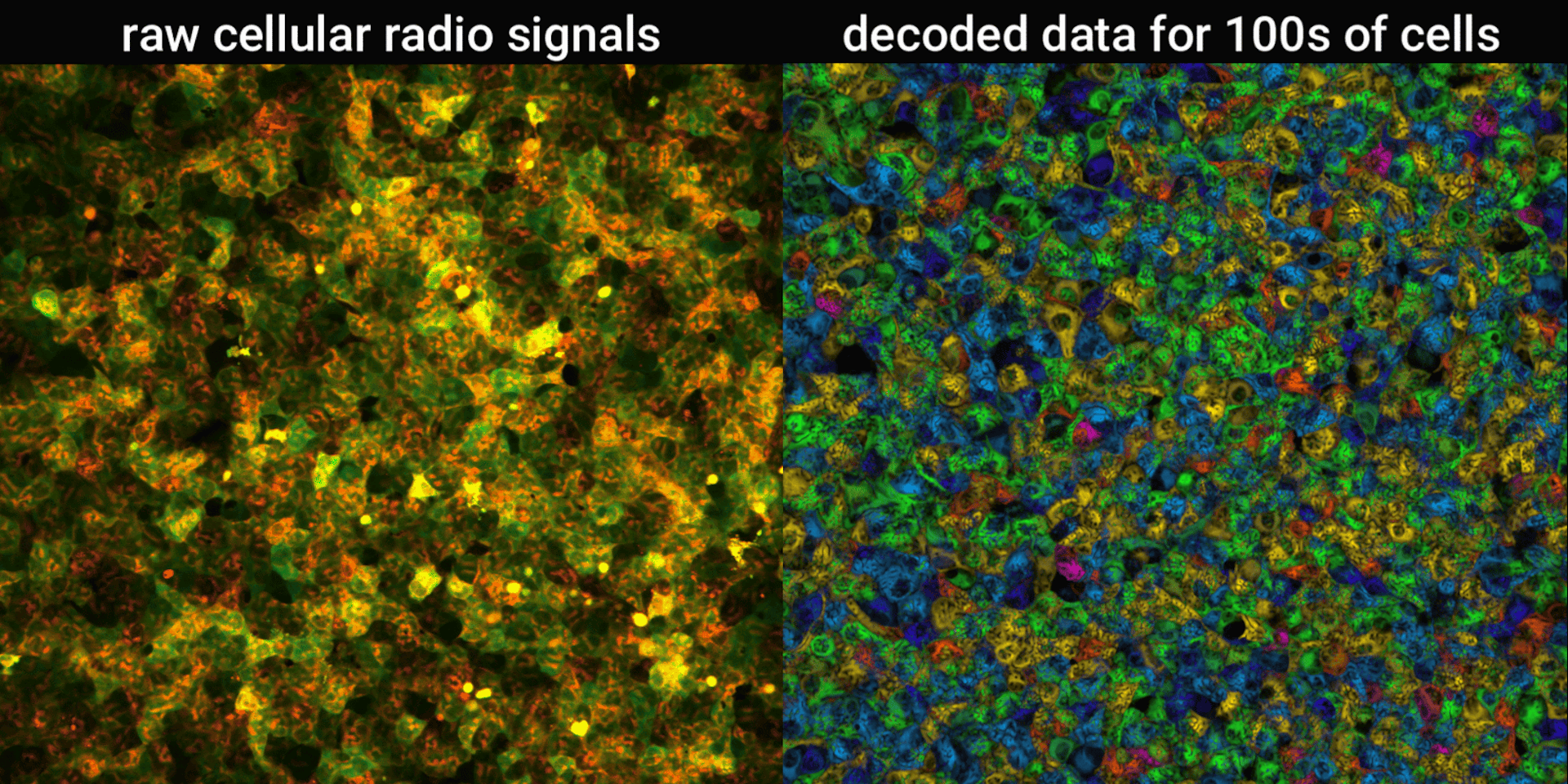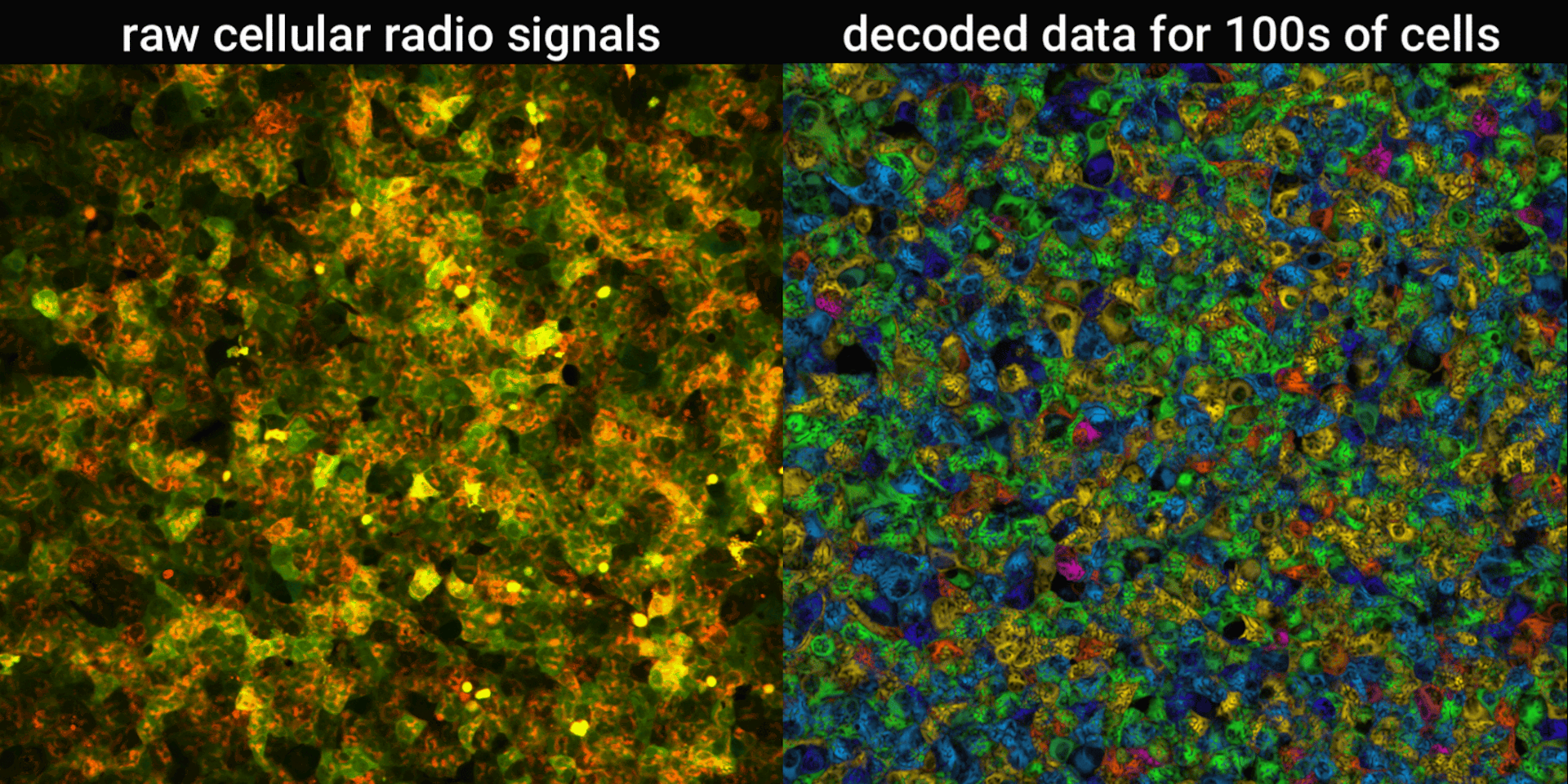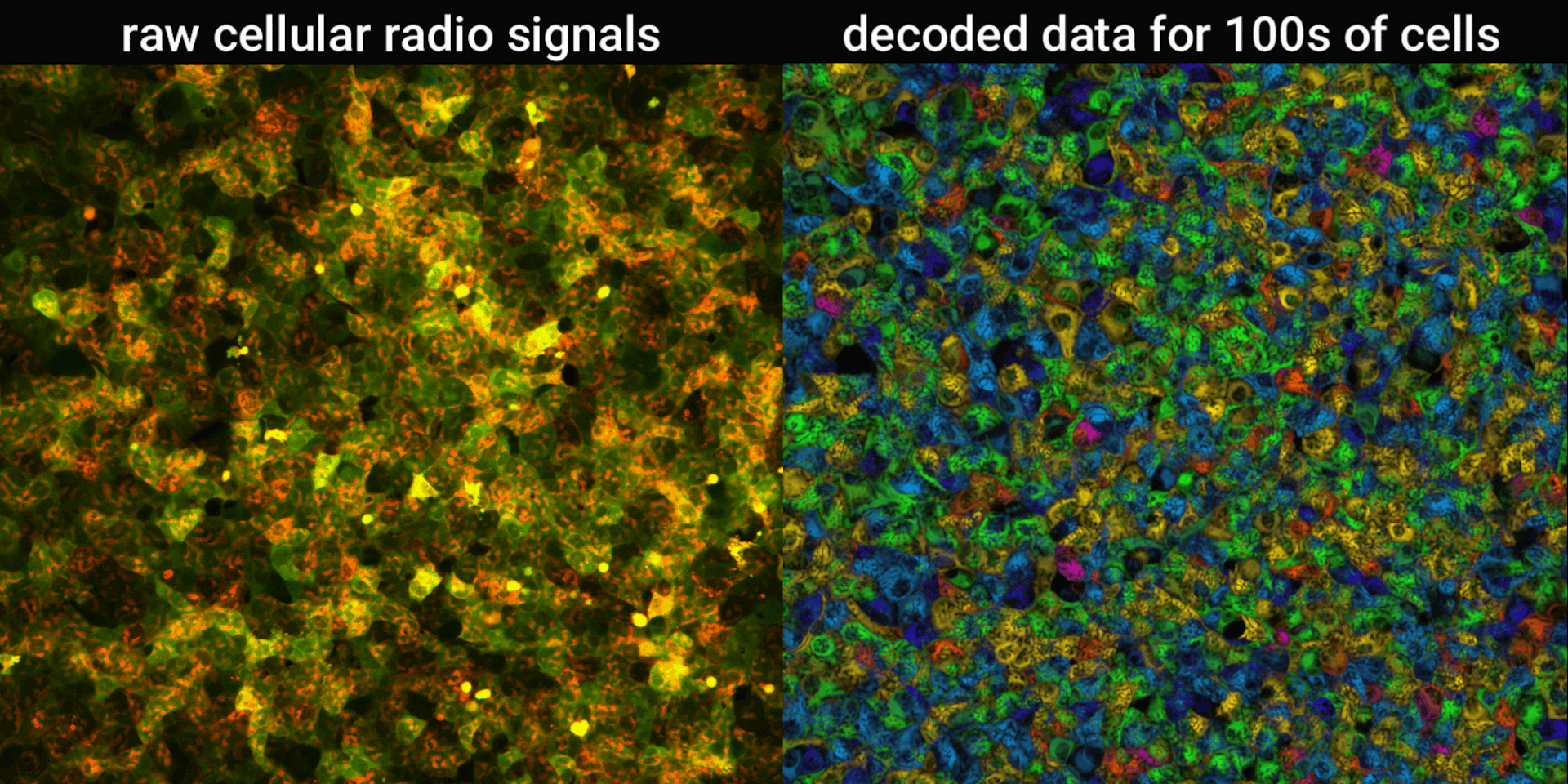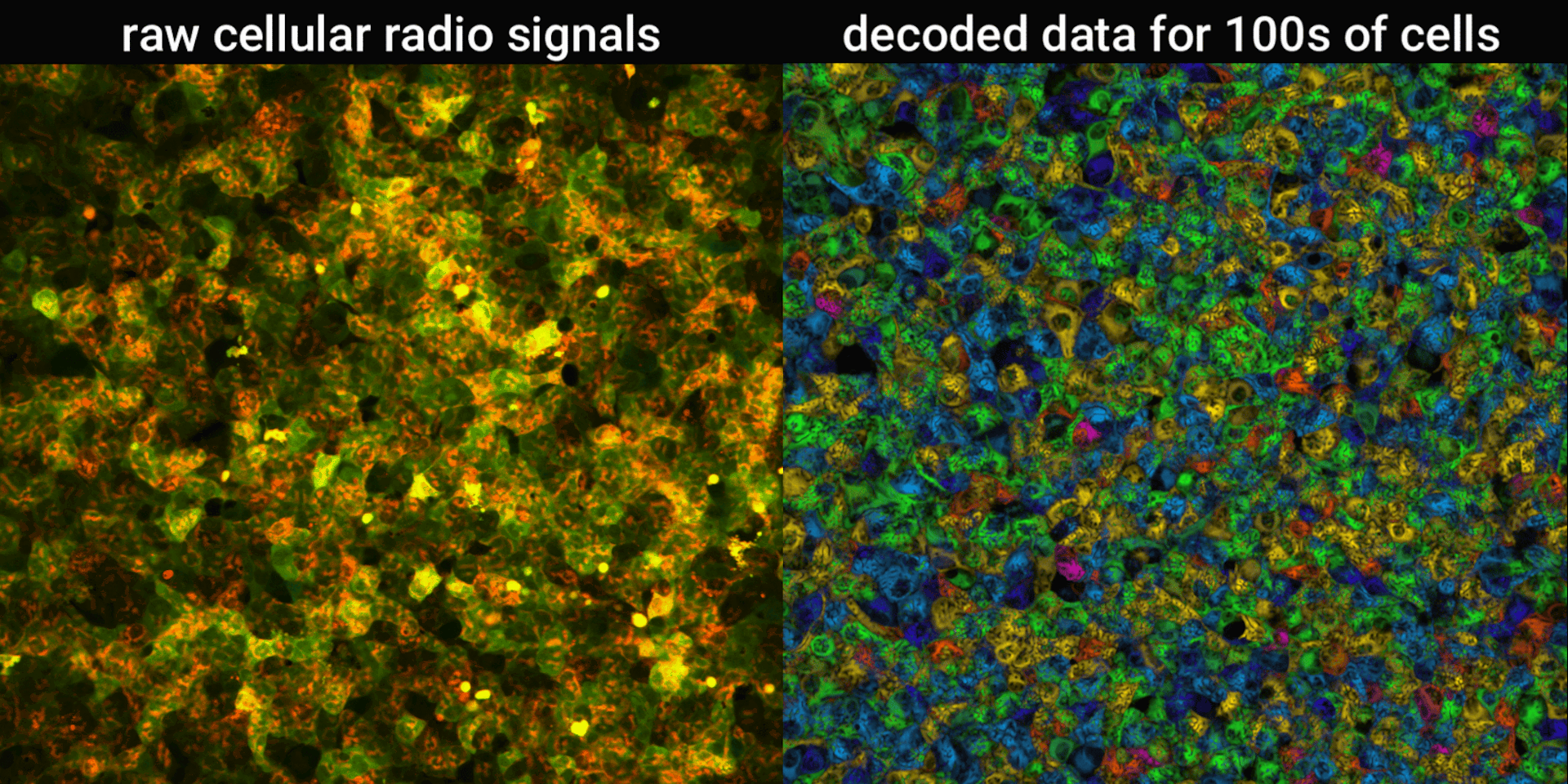
We engineered connections between MinDE protein waves and the activity of proteins responsible for key cellular processes. Now, the activity of these proteins trigger changes in the frequency or amplitude of the protein wave, just like an AM/FM radio. Using microscopes, we can detect and record the unique signals individual cells are broadcasting and then decode them to recover the dynamics of these cellular processes.
We have only begun to scratch the surface of how scientists can use protein waves to study cells. If the history of waves in technology is any indicator, their potential is vast.
This article is republished from The Conversation under a Creative Commons license. Read the original article.
![]()
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Women’s health cannot leave rare diseases behind
A physician living with lymphangioleiomyomatosis and a basic scientist explain why patient-driven, trial-ready research is essential to turning momentum into meaningful progress.

Life in four dimensions: When biology outpaces the brain
Nobel laureate Eric Betzig will discuss his research on information transfer in biology from proteins to organisms at the 2026 ASBMB Annual Meeting.

Fasting, fat and the molecular switches that keep us alive
Nutritional biochemist and JLR AE Sander Kersten has spent decades uncovering how the body adapts to fasting. His discoveries on lipid metabolism and gene regulation reveal how our ancient survival mechanisms may hold keys to modern metabolic health.

Redefining excellence to drive equity and innovation
Donita Brady will receive the ASBMB Ruth Kirschstein Award for Maximizing Access in Science at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Mining microbes for rare earth solutions
Joseph Cotruvo, Jr., will receive the ASBMB Mildred Cohn Young Investigator Award at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Fueling healthier aging, connecting metabolism stress and time
Biochemist Melanie McReynolds investigates how metabolism and stress shape the aging process. Her research on NAD+, a molecule central to cellular energy, reveals how maintaining its balance could promote healthier, longer lives.

