A sensor for fast, inexpensive on-site Ebola detection
Researchers are developing a new sensor that can detect Ebola in a single drop of blood and provides results in just an hour. With further development, the technology might also enable fast and inexpensive detection of other viruses, including the virus that causes COVID-19.
Ebola is one of the deadliest of all known viruses, killing up to 90% of those infected. Stopping its spread requires quickly detecting and isolating infected people. However, outbreaks tend to occur in remote areas of Africa, requiring blood tests to be transported to distant laboratories for analysis. This leads to significant delays in identifying a new outbreak.

Soma Banerjee, a visiting scientist in Marit Nilsen–Hamilton’s laboratory at Iowa State University, research associate in Ames National Laboratory and research scientist at Aptalogic Inc., will present the research at the American Society for Biochemistry and Molecular Biology annual meeting during the virtual Experimental Biology 2021 meeting, to be held April 27–30.
The new sensor is being developed by a research team led by Nilsen-Hamilton, who is also chief scientific officer of Aptalogic Inc., and Pranav Shotriya from Iowa State University. It involves a multidisciplinary effort that includes researchers with expertise in virology, bioinformatics, molecular biology and mechanical engineering from the University of Iowa, Iowa State University and the National Center for Biotechnology Information, part of National Institutes of Health’s National Library of Medicine.

The Ebola sensor is based on DNA aptamers, which are short, single-stranded DNA molecules that selectively bind to a specific target. To detect Ebola, the researchers identified aptamers that bind to Ebola virus soluble glycoprotein, a protein that appears in the blood of a person with Ebola before symptoms appear.
Current methods used to detect Ebola are based on analysis techniques that require laboratory facilities and trained individuals to perform the tests. Although alternative methods do exist, they tend to be difficult to read by personnel wearing protective gear or require special storage conditions.
“Our new sensor doesn’t require any special storage conditions,” said Banerjee. “This is an immense advantage because Ebola outbreaks occur frequently in remote areas where even electricity can be a luxury.”
So far, the researchers have shown that the aptamers they selected work well on a portable nanoporous alumin oxide sensor. They also found that the sensor can detect the Ebola glycoprotein in infected macaque blood serum, providing results that are comparable to the standard ELISA-based assay performed using Ebola antibodies.
“Once our device is fully optimized for detecting Ebola, we plan to develop a multiplexed version that can perform multiple tests and detect other viruses and microbes, all from one drop of blood,” said Banerjee. “We’re also using what we’ve learned so far to identify aptamers that could be used to detect COVID-19 and other similar viruses.”
The researchers are now working to further reduce the one-hour testing time without compromising the accuracy. They also want to make the sensor more user-friendly so that results can be read by personnel without any special training.
Banerjee will present the findings from 11:30–11:45 a.m. Friday, April 30 (abstract).
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
How sugars shape Marfan syndrome
Research from the University of Georgia shows that Marfan syndrome–associated fibrillin-1 mutations disrupt O glycosylation, revealing unexpected changes that may alter the protein's function in the extracellular matrix.

What’s in a diagnosis?
When Jessica Foglio’s son Ben was first diagnosed with cerebral palsy, the label didn’t feel right. Whole exome sequencing revealed a rare disorder called Salla disease. Now Jessica is building community and driving research for answers.

Peer through a window to the future of science
Aaron Hoskins of the University of Wisconsin–Madison and Sandra Gabelli of Merck, co-chairs of the 2026 ASBMB annual meeting, to be held March 7–10, explain how this gathering will inspire new ideas and drive progress in molecular life sciences.
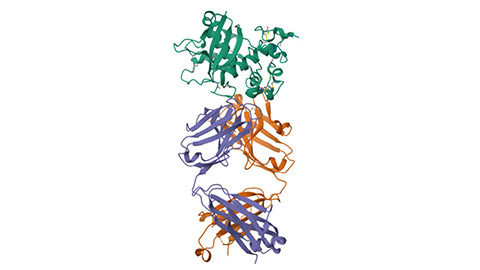
Glow-based assay sheds light on disease-causing mutations
University of Michigan researchers create a way to screen protein structure changes caused by mutations that may lead to new rare disease therapeutics.
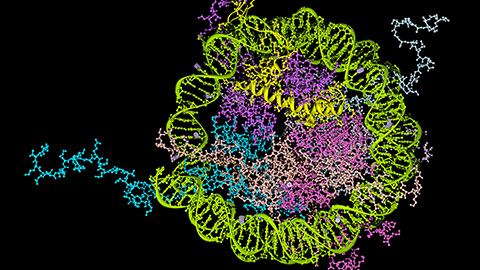
How signals shape DNA via gene regulation
A new chromatin isolation technique reveals how signaling pathways reshape DNA-bound proteins, offering insight into potential targets for precision therapies. Read more about this recent MCP paper.
A game changer in cancer kinase target profiling
A new phosphonate-tagging method improves kinase inhibitor profiling, revealing off-target effects and paving the way for safer, more precise cancer therapies tailored to individual patients. Read more about this recent MCP paper.

