Arginine tango
As a means to evade the host immune response, S. aureus uses an enzyme called oleate hydratase, or OhyA, to inactivate antimicrobial unsaturated fatty acids in the membrane that would otherwise inhibit bacterial growth. Research scientists at St. Jude Children’s Research Hospital reported today the structure and catalytic mechanism of OhyA.
Christopher Radka of St. Jude’s Department of Infectious Diseases describes the research during a presentation Tuesday at 2 p.m. EDT at the 2021 ASBMB Annual Meeting, held in conjunction with the Experimental Biology conference.
Radka and colleagues used X-ray crystallography to determine the structure of OhyA. Solving and evaluating multiple OhyA crystal structures highlighted a coordinated dance that occurs between key arginine residues and the unsaturated fatty acid substrate in the active site of the enzyme, a process facilitated by the nucleotide cofactor FAD.
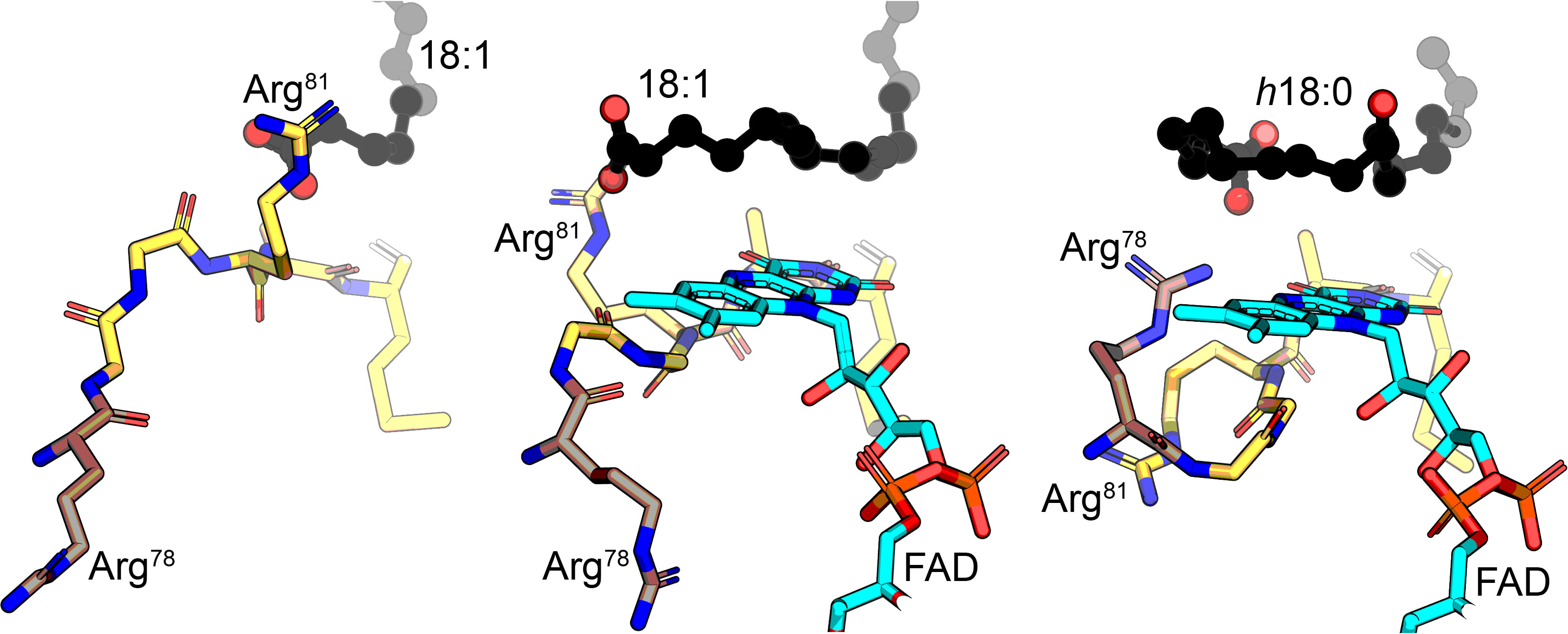
In this dance, the substrate is first guided into the binding tunnel by the oleate carbonyl of OhyA, then encounters its first arginine dance partner (Arg81) at the entrance of the active site. FAD binding then triggers the rotation of Arg81 that guides the fatty acid as it curls into the active site. After catalysis, a second arginine (Arg78) rotates behind the fatty acid carboxyl to release the hydroxylated product from the active site.
“What’s novel about the (active site) is how these conserved arginines guide the substrate through the donut-shaped active site,” Radka said. “Here, the arginines dance like two partners in a tango.”
This highly choreographed dance controls how the fatty acid substrate moves into and out of the active site. “In this coordinated tango at the active site, the FAD is the dramatic third character whose role is to come in and advance the dance so the chemistry can occur,” Radka said.
In this reaction, FAD remains oxidized and unconsumed. This quality is advantageous for industrial biotechnology research looking to use OhyA; FAD-dependent reactions often consume FADH2 and require continued starting product, which can be costly.
Future goals for this research include determining the structural elements required for S. aureus OhyA to remove antimicrobial fatty acids from the membrane.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
CRISPR epigenome editor offers potential gene therapies
Scientists from the University of California, Berkeley, created a system to modify the methylation patterns in neurons. They presented their findings at ASBMB 2025.

Finding a symphony among complex molecules
MOSAIC scholar Stanna Dorn uses total synthesis to recreate rare bacterial natural products with potential therapeutic applications.

E-cigarettes drive irreversible lung damage via free radicals
E-cigarettes are often thought to be safer because they lack many of the carcinogens found in tobacco cigarettes. However, scientists recently found that exposure to e-cigarette vapor can cause severe, irreversible lung damage.

Using DNA barcodes to capture local biodiversity
Undergraduate at the University of California, Santa Barbara, leads citizen science initiative to engage the public in DNA barcoding to catalog local biodiversity, fostering community involvement in science.

Targeting Toxoplasma parasites and their protein accomplices
Researchers identify that a Toxoplasma gondii enzyme drives parasite's survival. Read more about this recent study from the Journal of Lipid Research.

Scavenger protein receptor aids the transport of lipoproteins
Scientists elucidated how two major splice variants of scavenger receptors affect cellular localization in endothelial cells. Read more about this recent study from the Journal of Lipid Research.

