A versatile lipid second messenger
The phosphoinositide 3-kinase, or PI3K, signaling pathway has been studied intensively since its discovery in the mid-1980s and has been found to play a critical role in the regulation of normal cellular physiology. The pathway is dysregulated in pathophysiologies such as cancer and diabetes.
Much of this work focused on the lipid product of PI3K activity, PIP3, or PI(3,4,5)P3, derived from the phosphorylation of the membrane phosphoinositide PI(4,5)P2. PIP3 is a true lipid-derived second messenger: It is absent in unstimulated cells, it is synthesized rapidly at sites of PI3K activation at the plasma membrane, it is removed by the action of the lipid phosphatase and tumor suppressor PTEN, and it initiates PI3K signal relay by recruiting effector proteins such as the serine/threonine protein kinase Akt and downstream nodes such as the mTOR pathway.
By contrast, a separate lipid product of PI3K, PI(3,4)P2, remained largely out of the limelight. Recent studies point to an important role for this neglected lipid second messenger, with some surprises. Unlike PIP3, PI(3,4)P2 can be generated by more than one route of synthesis. 5' phosphoinositide phosphatases can dephosphorylate PIP3 to generate PI(3,4)P2. Alternatively, class II PI3-kinases can use PI4P as a substrate to generate PI(3,4)P2. And PI(3,4)P2 can be degraded by lipid phosphatases such as PTEN and INPP4B — tumor suppressors frequently inactivated in human cancers.
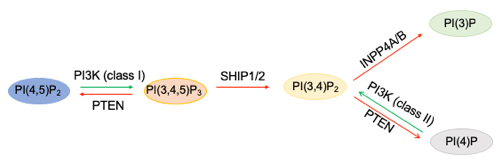 In phosphoinositide metabolism, PIP3, labeled PI(3,4,5)P3, is generated by phosphorylation of the precursor PI(4,5)P2 by the action of class I PI3-kinases, and the reverse reaction is generated by the phosphatase PTEN. SHIP1/2 lipid phosphatases dephosphorylate PIP3 to generate PI(3,4)P2, which is degraded further by INPP4A/B to PI3P. Class II PI3-kinases also can generate PI(3,4)P2 by phosphorylating PI4P. Alex Toker
In phosphoinositide metabolism, PIP3, labeled PI(3,4,5)P3, is generated by phosphorylation of the precursor PI(4,5)P2 by the action of class I PI3-kinases, and the reverse reaction is generated by the phosphatase PTEN. SHIP1/2 lipid phosphatases dephosphorylate PIP3 to generate PI(3,4)P2, which is degraded further by INPP4A/B to PI3P. Class II PI3-kinases also can generate PI(3,4)P2 by phosphorylating PI4P. Alex Toker
PI(3,4)P2–binding proteins, such as TAPP1/2 and lamellipodin, link PI3K/PI(3,4)P2 to cytoskeletal remodeling. However, many proteins with modular domains that directly bind to PIP3, including Akt, also bind to PI(3,4)P2. This has hampered identification of any potential unique signaling roles for PI(3,4)P2. Advanced high-resolution live cell imaging and microscopy show that PI(3,4)P2 is localized predominantly at early endosomes, although some plasma membrane localization is also evident. In growth factor signaling, this pool of PI(3,4)P2 seems to be generated predominantly by the action of phosphatases acting on PIP3. In contrast, PIP3 is localized exclusively at the plasma membrane, where class I PI3-kinases are activated.
These discrete intracellular pools of PI(3,4)P2 and PIP3 have remarkable consequences. In mammals, three Akt isoforms exist: Akt1, Akt2, and Akt3. Genetic ablation studies have shown that Akt isoforms play a non-redundant role both in normal development and in diseases such as cancer. Although a few isoform-specific substrates have been identified, of the 200-plus Akt substrate proteins identified, most are not unique to any one Akt. So how is specificity achieved? Recent research shows that Akt1 and Akt3 are activated specifically by the plasma membrane PIP3 pool, whereas Akt2 seems to be exclusively localized and activated by the endomembrane pool of PI(3,4)P2, at early endosomes and also early lysosomes. In the latter case, upon nutrient deprivation, class II PI3-kinases produce PI(3,4)P2 at the lysosome, leading to suppressed mTORC1 activity. Class II PI3-kinases also been have implicated in the formation of clathrin-mediated endocytic cups leading to vesicle fission, a process shown to be mediated by PI34P2.
A picture begins to emerge: Discrete pools of PIP3 and PI(3,4)P2 selectively activate Akt isoforms leading to presumably unique substrate phosphorylation patterns and downstream effects, although this has yet to be demonstrated rigorously. These observations begin to ascribe a critical role for PI(3,4)P2 in cell biology and also explain the roles of Akt isoforms in phenotypes associated with disease.
Questions remain about the intracellular localization of PI3K lipid second messengers and their effectors. The answers will come from continued advances in imaging technologies using live cells and tissues, giving a more complete picture of how specificity is achieved in this versatile lipid signaling pathway.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Targeting Toxoplasma parasites and their protein accomplices
Researchers identify that a Toxoplasma gondii enzyme drives parasite's survival. Read more about this recent study from the Journal of Lipid Research.

Scavenger protein receptor aids the transport of lipoproteins
Scientists elucidated how two major splice variants of scavenger receptors affect cellular localization in endothelial cells. Read more about this recent study from the Journal of Lipid Research.

Fat cells are a culprit in osteoporosis
Scientists reveal that lipid transfer from bone marrow adipocytes to osteoblasts impairs bone formation by downregulating osteogenic proteins and inducing ferroptosis. Read more about this recent study from the Journal of Lipid Research.

Unraveling oncogenesis: What makes cancer tick?
Learn about the ASBMB 2025 symposium on oncogenic hubs: chromatin regulatory and transcriptional complexes in cancer.

Exploring lipid metabolism: A journey through time and innovation
Recent lipid metabolism research has unveiled critical insights into lipid–protein interactions, offering potential therapeutic targets for metabolic and neurodegenerative diseases. Check out the latest in lipid science at the ASBMB annual meeting.
Melissa Moore to speak at ASBMB 2025
Richard Silverman and Melissa Moore are the featured speakers at the ASBMB annual meeting to be held April 12-15 in Chicago.


