Global analysis of coronavirus protein research reveals how countries respond to disease
In a new study, researchers examined how a country’s number of published 3D protein structures for coronaviruses, including the one responsible for COVID-19, correlated with its economic output and population. The findings reveal important insights into how different countries' research establishments respond to disease outbreaks and could be useful for planning responses to future pandemics.
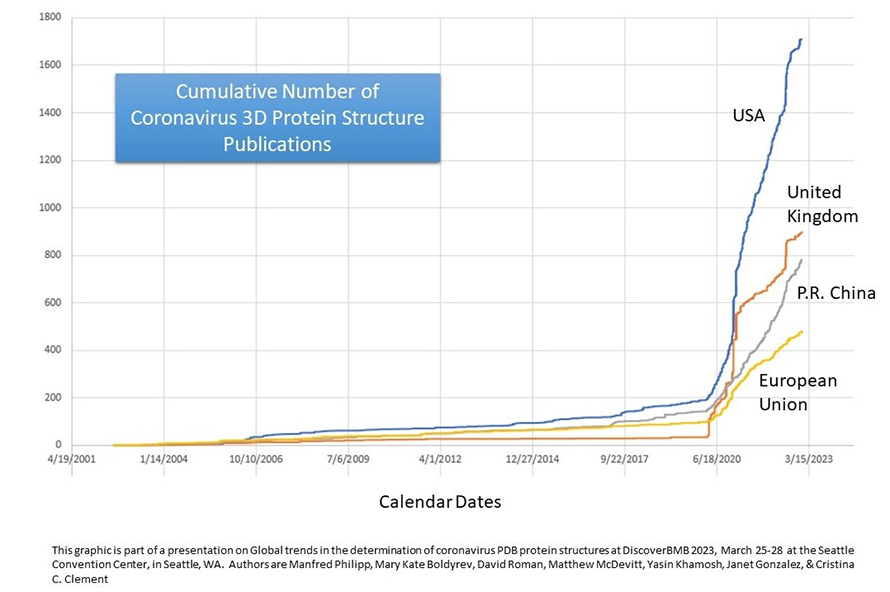
The study showed that countries with larger economies generated more 3D structure determinations for the protein components of coronaviruses, although there were many outliers. For example, some advanced and prosperous countries published few or no coronavirus structures while some countries strongly affected by COVID-19 published far more such structures than would be expected based on their populations and economies.
Manfred Philipp, professor emeritus at the City University of New York, will present the new research at Discover BMB, the annual meeting of the American Society for Biochemistry and Molecular Biology, March 25–28 in Seattle.
The 3D structures of proteins, such as those that make up coronaviruses, are determined using techniques such as X-ray crystallography and nuclear magnetic resonance. This information is then deposited into the Protein Data Bank (PDB) database so that it can be used to develop new drug treatments and new vaccines. Protein structure information is also key for understanding how viruses infect cells and interact with our immune system.
“We asked ourselves how different countries respond to disease, not in terms of treatment and medical care, but in terms of the research needed to study those diseases and to develop new treatments,” said Philipp. “Structure determinations provide a clear metric in terms of research response without any vagaries of interpretation.”
Although a great deal of research has examined levels of coronavirus research funding in various countries, most of this work has focused either on coronavirus-related patents or financial inputs. However, relating research to patents is complex, in part, because some nonprofits perform considerable amounts of research but rarely apply for patents while some patent-generating organizations don’t conduct a lot of research. Financial-based analyses don’t provide a complete picture because it is difficult to capture both public and private funds. Also, private research funds are not always publicly disclosed and some funds that are reported may be used for administrative and overhead costs instead of directly for research.
“Because we did not look at levels research funding, we didn’t have to figure out how much research funding was devoted to disease and how much to something else,” said Philipp. “Our metric only sees research successes and provides a clear indication of the amount of research applied to this topic alone.”
In the work, the researchers collected and cataloged 3D structure determinations for all proteins and nucleic acids that are components of coronaviruses. This included viruses from animal infections as well as those responsible for the 2003 SARS epidemic in Asia, the 2012 MERS epidemic in the Middle East, the worldwide COVID-19 pandemic and other coronaviruses that have been studied.
Of the approximately 40 countries the researchers examined, the United States, United Kingdom and China had the highest number of protein structures. They found that population size contributed very little to the number of coronavirus protein deposits while countries with a higher GDP did tend to deposit more protein structures. The number of confirmed COVID-19 cases was positively associated with the number of PDB deposits per country, especially in the U.S. Although South Africa and Brazil are not in the top 10 research-producing countries, they had a high number of COVID-19 cases and PDB deposits, suggesting that the high number of cases experienced in these countries influenced their research output.
“Our research on the country distribution of coronavirus-related structure determinations is intended to give research administrators in various countries the tools to redirect research funds in the most productive direction,” said Philipp. “For example, it can help countries with large research budgets but few if any structure determinations to redirect the focus of their research. This should not be the only such metric, but it is a key metric since it is independent of administrative and overhead costs.”
Next, the researchers plan to examine whether the number of structure publications for coronaviruses will continue to increase as the COVID-19 pandemic subsides and whether the countries that have led in depositing coronavirus structures into the PDB will continue to lead.
Manfred Philipp will present this research from 4:30–6:30 p.m. PDT on Sunday, March 26, in Exhibit Hall 4AB of the Seattle Convention Center (Poster Board No. 177) (abstract). Contact the media team for more information or to obtain a free press pass to attend the meeting.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Targeting Toxoplasma parasites and their protein accomplices
Researchers identify that a Toxoplasma gondii enzyme drives parasite's survival. Read more about this recent study from the Journal of Lipid Research.

Scavenger protein receptor aids the transport of lipoproteins
Scientists elucidated how two major splice variants of scavenger receptors affect cellular localization in endothelial cells. Read more about this recent study from the Journal of Lipid Research.

Fat cells are a culprit in osteoporosis
Scientists reveal that lipid transfer from bone marrow adipocytes to osteoblasts impairs bone formation by downregulating osteogenic proteins and inducing ferroptosis. Read more about this recent study from the Journal of Lipid Research.

Unraveling oncogenesis: What makes cancer tick?
Learn about the ASBMB 2025 symposium on oncogenic hubs: chromatin regulatory and transcriptional complexes in cancer.

Exploring lipid metabolism: A journey through time and innovation
Recent lipid metabolism research has unveiled critical insights into lipid–protein interactions, offering potential therapeutic targets for metabolic and neurodegenerative diseases. Check out the latest in lipid science at the ASBMB annual meeting.
Melissa Moore to speak at ASBMB 2025
Richard Silverman and Melissa Moore are the featured speakers at the ASBMB annual meeting to be held April 12-15 in Chicago.

