From the journals: JBC
With sulfur, life finds a way. Specific anti-cancer antibodies. Exercise-induced signaling cross talk. Read about articles on these topics recently published in the Journal of Biological Chemistry.
Life finds a way
Sulfur is one of the essential elements required for life. Plants and microbes often take up sulfur from inorganic sulfates in the environment using the sulfate assimilation pathway, or SAP, which reduces sulfates into sulfides and then synthesizes organosulfurs. In yeast, the SAP culminates in Met15-catalyzed biosynthesis of the amino acid homocysteine. Since its discovery, researchers have considered the gene encoding Met15 to be an essential marker for yeast growth in media lacking organosulfurs, and strains lacking Met15 have been used as the cornerstone of many genetic and genomic studies.

In their recent publication in the Journal of Biological Chemistry, S. Branden Van Oss, Saurin Bipin Parikh, Nelson Castilho Coehlo and colleagues at the University of Pittsburgh School of Medicine used structural and evolutionary modeling and genetic complementation experiments to show that the previously uncharacterized gene YLL058W encodes an alternative homocysteine synthase. They found that cells lacking Met15 still can assimilate inorganic sulfur and grow as long as excess sulfides are eliminated from the environment, indicating that Met15 is not essential for sulfur assimilation. In addition, the authors posit that the location of YLL058W near the unstable telomere region of the chromosome in all species that contain a homolog could indicate strong positive selective forces.
These results have implications for research on microbial and eukaryotic sulfur metabolism, including such aspects as the nutrient starvation stress response. This discovery also highlights how unknown variables can confound long-held assumptions.
Specific anti-cancer antibodies
The binding of programmed death-ligand 1, or PD-L1, to its receptor, programmed cell death protein 1, or PD-1, suppresses T cells and the immune system. Cancer cells frequently exploit this activity by overexpressing PD-L1 to evade immune activation; however, neutralizing monoclonal antibody therapy that targets PD-L1 has been effective in treating these cancers. Researchers recently have found that single-domain antibodies, such as nanobodies derived from camelids, may offer additional specificity and treatment options.
In a recent study in the Journal of Biological Chemistry, Tara Kang–Pettinger and colleagues at the University of Leicester used X-ray diffraction, NMR, AlphaFold and biolayer interferometry to solve a number of crystal structures of PD-L1 bound to nanobodies and characterize their binding interface. They found that the PD-1 binding surface on PD-L1 overlapped with another binding surface that recognizes CD80, a second receptor expressed on antigen-presenting cells that promotes a T cell anti-tumor response.
By comparing the binding sites of PD-1 and CD80, these researchers identified a binding region on PD-L1 specific for PD-1 and not for CD80 that could be bound by nanobodies. This binding permitted multiple simultaneous avenues to counteract PD-L1 overexpression and represents a step forward in the fight against cancer.
Exercise-induced signaling crosstalk
Physical inactivity and sedentary lifestyle are leading risk factors for obesity, Type 2 diabetes and heart diseases. Scientists know that the cytokine oncostatin M, or OSM, enhances insulin resistance in obesity through the phenotypic change of pro-inflammatory to anti-inflammatory macrophages when OSM is produced by adipocytes; however, researchers do not yet fully know what role OSM production plays in skeletal muscle after aerobic exercise.
Tadasuke Komori and colleagues at Wakayama Medical University in Japan reported in a recent article in the Journal of Biological Chemistry that OSM produced in the skeletal muscle after a single bout of aerobic exercise played a significant role in crosstalk between muscle and immune cells. Using OSM-deficient mice and direct intramuscular injection of OSM, they showed that OSM in the skeletal muscle was linked to the recruiting and accumulation of macrophages and neutrophils after exercise. Furthermore, they found that OSM induced the expression of a number of anti-inflammatory cytokines and markers.
These findings indicate that OSM is a novel myokine produced in muscle fibers and plays an important role in biological events such as the phenotypic determination of macrophages after aerobic exercise. This work could inform strategies for improving insulin sensitivity in muscle tissue.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
The data that did not fit
Brent Stockwell’s perseverance and work on the small molecule erastin led to the identification of ferroptosis, a regulated form of cell death with implications for cancer, neurodegeneration and infection.

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.
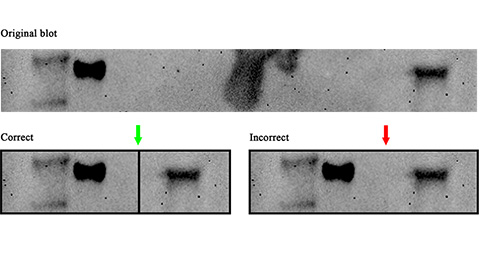
Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.
Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.
Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.
Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

