The myth of perfection
With the release of the imaging software Adobe Photoshop in the 1990s, “Photoshopping” entered the English lexicon. Like Google, Photoshop seamlessly has integrated itself into the scientific enterprise. Scientists use the software to tweak images and to generate publication-quality figures. It’s just so easy to create a blemish-free image. But there are guidelines to what is and isn’t acceptable to do with the software. There are a few simple rules to remember.
First, ask yourself whether any changes are needed. The best-case scenario is to be able to present your original, unaltered data in the figure. However, journal editors realize that sometimes the best case isn’t possible — an overly dark H&E stain or an overly bright Coomassie stain of a gel are two examples.
Once you’ve decided it’s appropriate and necessary to make changes, make sure your adjustments are linear. Most journals, including the journals published by the American Society for Biochemistry and Molecular Biology, require that adjustments be made uniformly to every pixel in the entire image. That means using the brightness and contrast functions in Photoshop is acceptable within reason, since these functions apply a linear adjustment to each pixel in the image. Also, go easy on moving the slider (see the figure). Overadjusting the brightness or contrast can hide background features, which is a misrepresentation of your data. Nonlinear adjustments include adjusting the gamma settings or using the “Curves” function in Photoshop. These actions are discouraged, since they do not apply changes equally to the pixels in the image. If these adjustments are used, then you must disclose their use in the figure legend.
 Aggressively overadjusting the brightness and/or contrast misrepresents the actual data that were obtained and can mask potential biologically relevant results.
Aggressively overadjusting the brightness and/or contrast misrepresents the actual data that were obtained and can mask potential biologically relevant results.
Speaking of data misrepresentation, specifically enhancing, removing or obscuring features would fall into this category. Worried that a faint band won’t support your conclusions? Bothered by the cell debris in the corner of your image? Concerned that the reviewers may say that the co-localization or the co-immunoprecipitation isn’t strong enough? The temptation to enhance or remove these features is real, but this type of manipulation falls into the misconduct category and could have serious consequences.
The final image should look like your original data, warts and all. You always should inspect your final figure and ask yourself if it is a true representation of the original capture or image. If your answer is no (or kind of), you should re-evaluate your figure.
Practically speaking, if any of these issues are discovered during the review of your paper or even after it is published, they could delay publication of your article, result in a correction, or even end in a retraction. More importantly, these issues go deeper and speak about the reproducibility of the work and your integrity as a scientist. Other researchers will not be able to replicate the results shown in your article if some of the data have been enhanced or hidden selectively. Presenting your data in a transparent manner ensures that you have done your due diligence.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
The data that did not fit
Brent Stockwell’s perseverance and work on the small molecule erastin led to the identification of ferroptosis, a regulated form of cell death with implications for cancer, neurodegeneration and infection.

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.
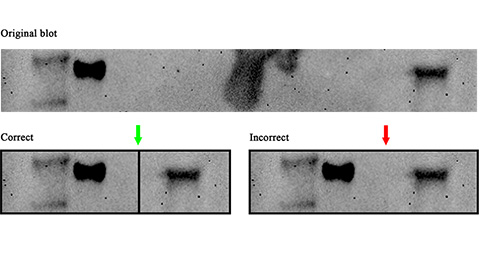
Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.
Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.
Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.
Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

