The odyssey of autophagy
If you’re a problematic protein, pathogen or plaque-precursor peptide, the encroaching of a Pac-Man-like autophagosome is the beginning of the end. Signaled by stress or a scarcity of nutrients, a double-layered membrane begins to form in a cell’s cytoplasm. After expanding, this autophagosome surrounds the undesirable cellular component, closes it off and joins membranes with a destructive sack of enzymes known as the lysosome. Once the undesirable component has been broken down, the lysosome opens up, releasing the constituent amino acids, lipids and other cellular building blocks back into the cytoplasm for reuse.
 Yoshinori Ohsumi
Yoshinori Ohsumi
As crucial as this process is for cell survival, it wasn’t well understood until 1993, when an associate professor at the University of Tokyo identified the genes responsible for inducing autophagy in yeast cells. The professor’s discovery reverberated through nearly all domains of cell biology over the coming decades and revolutionized biomedical research.
This October, the Nobel Committee for physiology or medicine honored that researcher, Yoshinori Ohsumi at the Tokyo Institute of Technology’s Frontier Research Center, for his pioneering work in discovering the molecular mechanisms for autophagy.
Ohsumi’s “discoveries opened the path to understanding the fundamental importance of autophagy in many physiological processes,” said the Nobel Assembly at Karolinska Institutet in Sweden in a press release.
Nobel laureate Randy Schekman at the University of California, Berkeley, says, “This pathway, as we know in mammalian cells, now touches every corner of the cell in terms of metabolism, regulation, control of viral and bacterial infection, and even the tumor potential of transformed cells,” adding that the recognition of Ohsumi’s work was well-deserved. Ohsumi is a member of the American Society for Biochemistry and Molecular Biology, which publishes ASBMB Today.
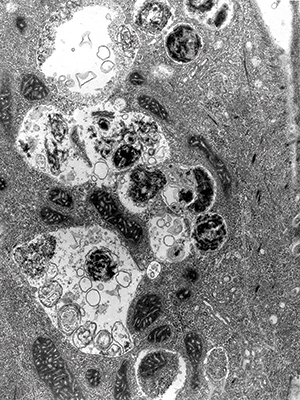 Electron micrograph shows mature autophagosomes engulfing intracellular components.IMAGE PROVIDED BY SHARON TOOZE
Electron micrograph shows mature autophagosomes engulfing intracellular components.IMAGE PROVIDED BY SHARON TOOZE
Rediscovery
The word “autophagy,” which means “self-eating” based on its Greek roots, was coined by the Nobel laureate Christian de Duve of The Rockefeller University and Université Catholique de Louvain in Belgium. After reading a paper by Thomas Ashford and Keith Porter about the presence of membranous sacs containing degraded cellular components in mouse kidney cells, de Duve began investigating the structures.
As it turns out, Porter and Ashford had misidentified the autophagosomes as nascent lysosomes rather than as transient organelles that fused with the lysosomes. De Duve had helped discover lysosomes in the 1950s. After several years of investigation in which de Duve found that autophagosomes increased in number as cellular degeneration increased, he named the process autophagy at a symposium on lysosomes in 1963.
However, for the next three decades, the study of autophagy was almost entirely observational and quite difficult. Unlike many other cellular components, the autophagosomes are short-lived, only existing for 10 to 20 minutes at a time. For comparison, the lysosomes with which autophagosomes fuse exist in the cytoplasm for several hours.
Enter Ohsumi. Ohsumi was born in 1945 in Fukuoka, Japan. After earning a bachelor’s degree from the University of Tokyo in 1967 and a Ph.D. from the same institution in 1974, Ohsumi began exploring cellular processes in yeast as a postdoctoral fellow at The Rockefeller University in New York City. At Rockefeller, Ohsumi, who was working with mice for his experiments on fertilization, got into working with yeast as a model organism (see “Fortuitous yeast”).
In 1977, Ohsumi returned to the University of Tokyo as a junior professor in the laboratory of Yasuhiro Anraku and began exploring the functions of yeast vacuoles, organelles that are homologous to human lysosomes.
Ohsumi continued investigating the vacuoles’ active transport systems and lytic functions for the next 11 years, opening his own small laboratory at the University of Tokyo in 1988. Five years later, Ohsumi reported in the journal FEBS Letters the discovery of 15 genes responsible for autophagy in yeast, which would come to be known as the ATG genes.
By growing yeast mutants that lacked vacuolar proteases on a nutrient-restricted medium, Ohsumi created a system in which autophagic bodies accumulated in the vacuole. After subjecting these yeast mutants to a process that generated random genetic mutations, he eventually found a mutant in which the autophagic bodies didn’t accumulate. The gene he’d knocked out, soon to be deemed “autophagy-related gene 1,” or ATG1, was essential to their formation.
“Yoshinori is really one of the founders of the autophagy field as we know it,” says Matthias Peter at the Swiss Federal Institute of Technology in Zurich. Peter says that Ohsumi’s significant contribution was that he didn’t just observe the pathway by microscopy but also “used an elegant genetic screen to identify molecular components that are required for autophagy. He laid down the genetic foundation of this pathway.”
Ohsumi and his laboratory members identified the critical genes responsible for each step of the autophagy pathway. Those discoveries allowed for the full role of autophagosomes to be mapped through the use of genetic screens. This would allow Ohsumi’s laboratory and others to investigate the ramifications of each step in the clearance pathway at the biochemical level.
After the breakthrough in 1993, when Ohsumi and colleagues published the FEBS Letters paper describing all 15 ATG genes, Ohsumi’s laboratory members set out to characterize the functions of each step of the pathway. Noboru Mizushuma was a postdoctoral researcher in Ohsumi’s laboratory who discovered that the Atg12 protein formed an essential trimolecular complex with Atg5 and Atg16. This complex promotes the conjugation of the protein Atg8 to the phospholipid phosphatidylethanolamine. The conjugated phospholipid then drives the coalescing autophagosomal membrane to elongate around intercellular components targeted for degradation and to fuse together, sealing the target off on its way to the vacuole. Linking these pieces together allowed Ohsumi’s team to present the entire pathway in several papers, including two papers in Nature in 1998 and 2000.
Working with Ohsumi “undoubtedly broadened my scientific view,” says Mizushima, who is currently at the University of Tokyo. “The most important lesson I learned during my time in Dr. Ohsumi’s lab is that researchers should have the courage and determination to pursue research directions and answer questions that they believe to be important irrespective of current trends and apparent usefulness.”
In a few years, Ohsumi and colleagues identified the mammalian homologues of the yeast ATG genes, resulting in a cascade of applications throughout cell biology.
“This long-anticipated and extremely well-deserved prize reminds us that the best way to make important discoveries is often to ask a simple question about an interesting phenomenon, pick the right model organism in which that question can be approached genetically and biochemically, and let the grand unity of biology do the rest,” says Gregory Petsko at Cornell University, a former ASBMB president.
Another former ASBMB president, Suzanne Pfeffer at Stanford University, agrees. “Dr. Ohsumi is a wonderful choice for this award,” she says. “He is a dear and humble man who used the power of yeast genetics combined with microscopy and biochemistry to work out the entire, unexpected pathway of autophagy that is highly conserved from yeast to humans.”
Fortuitous yeast
The research that ended up transforming so much of modern cell biology was born out of a period of isolation in Ohsumi’s career. While investigating the in vitro fertilization of mouse eggs as a postdoctoral researcher at The Rockefeller University in 1975, Ohsumi crossed paths with Michal Jazwinski, an incoming postdoctoral researcher.
According to Jazwinski, Ohsumi’s initial work in the lab of Gerald Edelman involved studying the processes involved in the chromatin decondensation of sperm cells.
“It was a fairly large laboratory, dealing with signals at the cell surface and interactions between cells,” says Jazwinski, now at Tulane University. While Ohsumi’s work involved cell–cell interactions, “it was a project that was off on the periphery. He was the only one that was looking at that phenomenon.”
As Jazwinski’s own research with signal transduction in lymphocytes began steering him toward investigating cell division in yeast, he says, he found himself venturing away from the rest of the laboratory.
“We were both isolated, which led to us gravitating toward each other,” says Jazwinski, who describes Ohsumi as an easygoing person. This led to Ohsumi beginning to work with Jazwinski on studying DNA duplication within the yeast cells. “We started interacting more. It was fun, because we had a way of being able to exchange ideas about very basic concepts in biology and practical things with experiments.”
Jazwinski started using yeast for his experiments. Ohsumi, recalls Jazwinski, “observed what I was doing and became interested in what I was doing.” That seed of interest, it seems, grew into a lifelong love of the intricacies of yeasts that Ohsumi still is pursuing to this day.
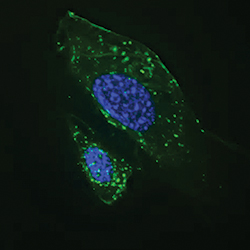 Autophagosomes (green) become activated in starved cells.IMAGE PROVIDED BY JAN PETRASEK AND BETH LEVINE
Autophagosomes (green) become activated in starved cells.IMAGE PROVIDED BY JAN PETRASEK AND BETH LEVINE
Autophagy’s many facets
Autophagy has been observed in many life forms, not just yeast and humans. According to Beth Levine at the University of Texas Southwestern Medical Center at Dallas, autophagy’s ubiquity across eukaryotes might be due to its early evolutionary role in facilitating eukaryotic cells’ ability to tolerate a variety of environments.
“If an organism can degrade and recycle its internal contents to survive short-term periods of starvation, this has tremendous advantages for evolution. The organism can now migrate to other environments and undergo selective mutations that permit it to adapt to other environments,” says Levine. “This facilitates evolutionary diversity.”
As a self-eating process, autophagy plays a significant role in intercellular nutrient regulation and homeostasis. “Autophagy is fundamental for nutrient homeostasis, sensing amino acid levels and nitrogen levels in both yeast and mammals,” says Sharon Tooze at the Francis Crick Institute in the U.K. “It’s not just a pathway for getting rid of garbage. It’s also a pathway that keeps cells in a nutrient-stable condition.”
Iron regulation, says Tooze, is one of the most essential pathways that autophagy recently has been found to control. The autophagy protein WIPI4 is involved in the turnover of ferritin, a protein that stores iron in a nontoxic form. When defects arise in WIPI4, iron can accumulate toxically in the brain, causing a condition called “neurodegeneration with brain iron accumulation disorder.”
Neurodegeneration from iron accumulation isn’t the only effect autophagy has on physiology when it goes awry. Researchers working with neurodegenerative disorders say one of the frontiers in autophagy is the ability to modulate the process selectively in certain parts of the body.
“Normally you activate autophagy every day, in all the cells in your body,” says Ana Maria Cuervo at the Institute for Aging Research of the Albert Einstein College of Medicine. “But the level of activation changes with age. In some organs, it cannot reactivate properly.”
Within Parkinson’s, Alzheimer’s and Huntington’s diseases, says Cuervo, there is a primary defect in autophagy. “Autophagy usually contributes to eliminate (excess) proteins. That’s why you don’t see Alzheimer’s patients who are 20 years old,” says Cuervo.
As autophagy starts declining with age, Cuervo explains, dysfunctional proteins such as beta-amyloid peptides or alpha-synuclein oligomers start accumulating and clogging the autophagy system, creating a vicious circle. “I think that the challenge right now is to try to understand in which diseases the changes in autophagy are the cause of the disease, and in which ones these changes that we see in autophagy are reactive to the disease itself,” she says.
While it would be ideal in these instances to ramp up autophagy in neurons, “something that we still don’t have is a drug that can very selectively only modulate autophagy,” says Cuervo.
Beyond self-eating, another crucial role of autophagy is its ability to remove intercellular pathogens, which include viruses, bacteria and parasites. This aspect of autophagy is known as xenophagy. One of the many research foci of Levine’s laboratory is understanding how the cell’s self-eating pathway can be used to degrade foreign material.
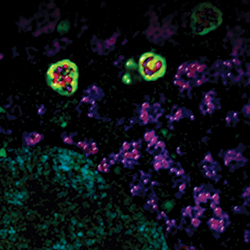 Autophagosomes (green) surround mitochondria (red).IMAGE PROVIDED BY RHEA SUMPTER AND BETH LEVINE
Autophagosomes (green) surround mitochondria (red).IMAGE PROVIDED BY RHEA SUMPTER AND BETH LEVINE
Disabling the autophagosome’s ability to identify foreign objects is a common pathogenic strategy. One example is herpes simplex virus, which generates viral virulence proteins that block the function of the Beclin-1 autophagy protein in neurons. This action causes significant cell damage, ultimately resulting in fatal encephalitis in mice.
By binding to the Beclin-1 protein in neurons, herpes simplex virus can block the cells’ autophagy function. Such is the strategy of herpes simplex virus as well as other viruses that target the central nervous system.
For a bacterium like Salmonella, says Levine, “the ability of the autophagic machinery to degrade the microbe basically determines whether the bacterium is or is not an intercellular pathogen.” In humans, Salmonella are capable of invading epithelial cells but usually are caught and destroyed by autophagosomes.
In roundworms, Salmonella typically replicates in the lumen of the intestine rather than the epithelium. When the autophagy machinery is knocked out in worms, Salmonella spreads through the epithelium unchecked, killing the worms.
Since 2004, there have been numerous studies from labs worldwide demonstrating that in higher eukaryotic organisms both the autophagy pathway and autophagy proteins play a crucial role in many different aspects of immunity, says Levine.
In good company
The Nobel Committee for physiology or medicine cited a 2007 paper from the Journal of Biological Chemistry among their reference materials for the prize. In the paper, Ohsumi and his colleagues demonstrated that a conjugate between the Atg12 and Atg5 proteins strongly enhances the formation of another conjugate, Atg8-phosphatidylethanolamine. Both of the ubiquitin-like conjugation systems are essential for autophagosome formation, but their relationship to one another was previously unclear.
Dramatic change
From cellular garbage can to nutrient-recycling, plaque-and-pathogen-punishing Roomba, the view of the autophagosomes’ role in cellular functions has changed dramatically since Ohsumi’s landmark discovery more than two decades ago. By teasing out the function of a pathway that most of his peers had ignored, Ohsumi gave cell biologists the tools they needed to explore deeply the mechanisms behind what is swallowed and what is spared.
Experts are excited about the future of this once-obscure niche of cell biology. “There are still many questions regarding regulation from these autophagosomes,” says Cuervo. “From the point of view of cell biology, that is the beauty of such a fundamental process.”
Ohsumi too marvels at the beauty of autophagy but pays tribute to the hard work of his fellow scientists. “As a scientist, there is no greater satisfaction that having your work recognized, and as the highest scientific honor, the Nobel Prize is very significant,” says Ohsumi. “Our discovery of the basic process of autophagy has opened up whole new areas of research. The selective degradation of organelles, protein aggregates and invading bacteria, for example, are physiologically very important phenomena that are now being rapidly uncovered by pioneering scientists all over the world. It is also becoming clear that the medical applications of our basic research in the treatment of cancers and other diseases are ever closer to realization, which is very exciting. I would therefore like to express my gratitude to researchers from all over the world who have shown interest in our work, made important contributions to the field and joined us on this exciting journey of discovery. The growth of the field would not have been possible without their efforts.”
The image on the top of this page, which is the same as this month's cover, shows how lipids accumulate in cells when autophagy fails. Low lipid levels are seen in blue; high lipid levels are in red. The image was done by Susmita Kaushik at the Albert Einstein College of Medicine.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
From the Journals: JBC
How cells recover from stress. Cancer cells need cysteine to proliferate. Method to make small membrane proteins. Read about papers on these topics recently published in the Journal of Biological Chemistry.

ASBMB names 2025 JBC/Tabor Award winners
The six awardees are first authors of outstanding papers published in 2024 in the Journal of Biological Chemistry.
Pan-kinase inhibitor for head and neck cancer enters clinical trials
A drug targeting the scaffolding function of multiple related kinases halts tumor progression.

Sweet secrets of sperm glycosylation
Scientists from Utrecht University uncover similar glycosylation patterns in sperm from bulls, boars and humans, distinct from those found in blood across species. These findings may improve IVF and farming techniques.
From the Journals: JLR
Promising therapeutic candidate for steatosis. Unique lipid profiles in glycogen storage disease. Microglial lactic acid mediates neuroinflammation. Read about these recent papers.

Meet Robert Helsley
The Journal of Lipid Research junior associate editor studies chronic liver disease and was the first in his family to attend college.

