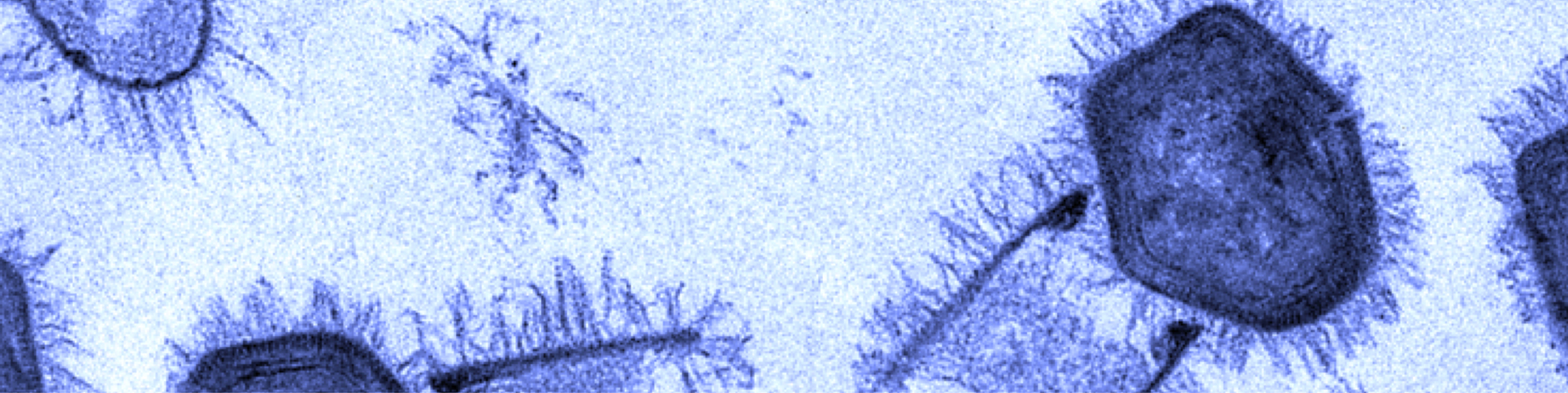
Giant gene thieves
If the words “giant viruses” strike fear — or nihilistic delight — you need not worry. The vast majority of the pseudo-lifeforms want nothing to do with humans, preferring to prey instead on cyanobacteria, phytoplankton, and protists and prokaryotes writ large. The corpulent cocci-catchers are of interest, however, to a close-knit community of virus hunters, geneticists and structural biologists.

One of those structural biologists, Chuan Xiao, who goes by River among colleagues and in correspondence, is an associate professor at the University of Texas at El Paso, where he co-directs the university’s cryo-electron microscopy facility. Xiao recently received a $1.75 million grant from the National Institutes of Health to continue characterizing the processes by which a giant virus, cafeteria roenbergensis virus, or CroV, assembles its protein shell.
Xiao’s analysis of CroV is made possible by a 14-foot-tall electron microscope housed in an interference-minimizing room — a dedicated piece of equipment that, while not rare, carried a $1.8 million price tag for the department and demands intense computing power to render images of viruses that are more than three times larger than HIV or influenza virions.
“It took me almost two years about — 3 million CPU hours at the supercomputer center — to get the reconstruction (of CroV’s membrane) to 14 Angstrom definition,” Xiao said. “You almost can see the 14 atoms lined up together — that’s how accurately we can put all these pieces together.”

While some of these viruses have potential medical applications — the methods CroV uses to assemble its protein shell, or capsid, are similar to those used by influenza viruses — many more play a quiet role in bringing balance to aquatic ecosystems.
Massive missives
When Bradfordcoccus first was isolated from the amoebae that had caused a pneumonia outbreak in Bradford, England, in 1992, it refused to replicate in the lab. This response confounded the researchers at the Université de la Méditerranée in Marseille, France — Bradfordcoccus was the size of a small bacterium, and it responded to Gram staining but not to amplification of 16S ribosomal RNA, a slowly evolving sequence used to reconstruct prokaryotic evolutionary trees.
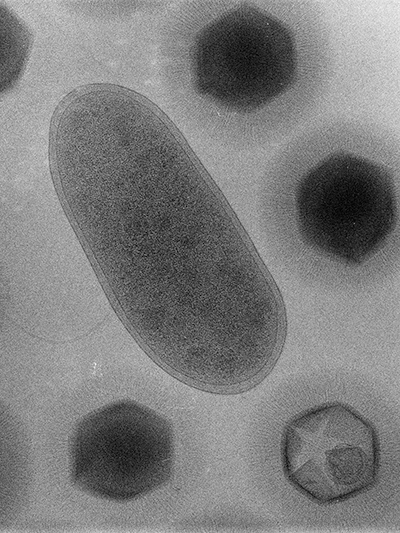
It took several years for the French microbiologists Didier Raoult and Bernard La Scola to recognize that Bradfordococcus was, in fact, a virus nearly as large as the amoeba Acanthamoeba polyphaga that it was hunting. In a paper published in the journal Science in 2003, La Scola, Raoult and their colleagues renamed the pathogen Acanthamoeba polyphaga mimivirus, and “mimivirus” — short for “mimicking microbe” — stuck.
Some of Raoult’s samples made their way to the lab of the late Michael Rossmann, a structural biologist and X-ray crystallography pioneer who led the first team to map the common cold on an atomic level, at Purdue University.
At the time, Xiao was a graduate student in Rossmann’s lab working with picornaviruses, a viral family that includes Rhinovirus, Poliovirus and Hepatovirus. He immediately was drawn to the challenge presented by the new, massive quarry.
“Cryo-EM was just beginning to become a powerful tool,” Xiao said. “Mostly we were working on smaller viruses (because) we still couldn’t get high resolution.”
In cryo-EM, researchers flash-freeze molecules and microorganisms in an aqueous environment and then fire electrons at them with a transmission electron microscope, gathering data about a sample from the electrons that pass through it. This gets around the difficulty of crystallizing samples but results in a lower-resolution image.
When the mimivirus samples arrived, Xiao was one of the only students capable of both operating the electron microscope and writing programs to compensate for the limitations of the computer’s encoded memory running up against the sheer size of the micrographs. Xiao, who had almost finished his thesis, quickly wrapped up his work on picornaviruses and stayed on as a postdoctoral fellow in the lab.
In autumn 2005, just months after he received his Ph.D., Xiao’s cryo-EM description of mimivirus at the level of 70 angstroms appeared in the Journal of Molecular Biology. For Xiao, the paper marked the beginning of a nearly three-year struggle to reconstruct the virus’ membrane, which is dotted with asymmetrical, rodlike fibers.

“It’s very hard to work with, because those fibers don’t follow any symmetry. They just add noise to your images,” he said. “I spent close to two and a half years trying to break through it, because at that time, we didn’t have an electron detector.”
The recent explosion of cryo-EM facilities has been largely thanks to the development of direct electron detectors, which allow for digital renderings of cryogenic samples.
“Now, we have cameras that can essentially detect electrons and count electrons,” said Thomas Klose, the technical director at the Purdue Cryo-EM Facility and a frequent collaborator with Xiao. “So we can recover much, much more of the theoretical signal that one would expect in an image. And that makes it easier to see things, and the contrast of your image improves greatly.”
But when Xiao was a postdoc, images created by the scattering of electrons off a sample’s surface were recorded on film that needed to be developed in a darkroom. His portfolio ran into the thousands.
“I collected almost 3,000 films. So just think about developing 3,000 films in a darkroom,” he said. “Every session of cryo-EM, we could only take about 128 films maximum … we did a reconstruction, but never were able to improve resolution.”
In 2008, Xiao took a job in the University of Texas at El Paso’s emerging structural biology program. Within two years, a grant from the National Science Foundation would fund the towering modern microscope Xiao and his colleagues use today.
A lost domain?
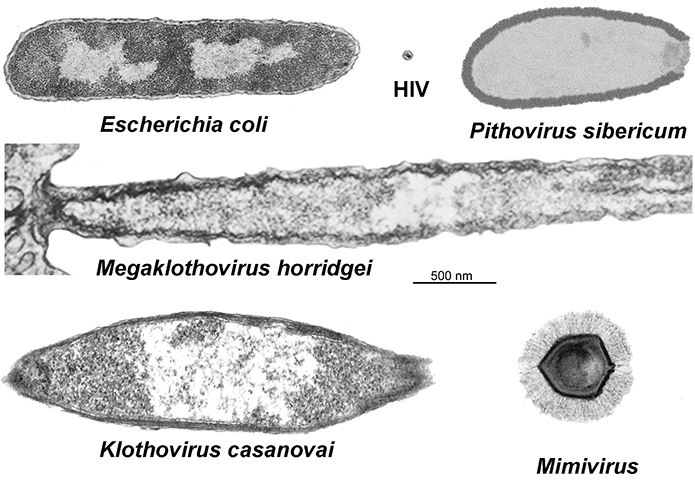
While mimivirus was the first identified nucleocytoplasmic large DNA virus, or NCLDV, it wasn’t alone for long. Giant viruses began cropping up anywhere wet — off the coast of Chile, in a 30,000-year-old Siberian ice core, inside an amoeba in the contact lens of a woman with keratitis — all of them bulked up with almost enough transcriptional machinery to exist outside of a host.
Because these massive genomic caches had previously been seen only in eukaryotic cells, their presence left microbiologists and geneticists unsure whether the viruses had evolved from smaller viruses that had scooped up genes from hosts or from cellular ancestors trading lives of full-time protein synthesis and ribosomal activity to live off the ATP of others.

According to Matthias Fischer, an environmental microbiologist at the Max Planck Institute for Medical Research in Heidelberg, Germany, who specializes in gigantic viruses and provides samples for structural biologists like Xiao, the latter hypothesis, suggesting a lost fourth domain of life outside of Archaea, Bacteria and Eukarya, was tantalizing for a time.
“There were two opposite hypotheses,” Fischer said. “One was that they reduced from a cellular ancestor and became viral, and that’s why they’re so large, because they retained a lot of the genes they had previously.”
Comparative genomics, however, appears to have provided an answer in the form of a small number of genes, including those used to make viral capsids and the DNA polymerase essential for viral replication, that are conserved across all known giant viruses.
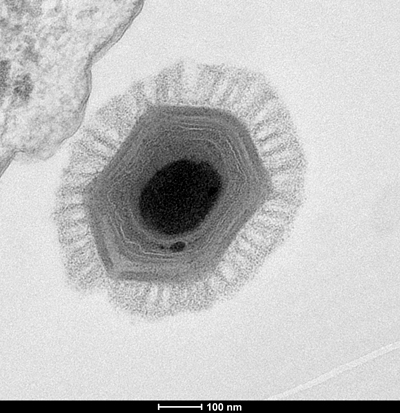
“It’s sometimes tricky to infer a common ancestry based on less than a percent of your genome,” Fischer said. “But these genes are so-called core genes, and they are present in so many different viruses, because they’re very important.”
The remaining 99% of giant virus genomes, which sometimes can include virophages, could be accounted for by the mechanisms the viruses use to cycle through genes, something Fischer likens to a “genomic accordion.”
“There’s a hypothesis called the genomic accordion, where these giant viruses can duplicate genes very rapidly to select for the best adapted version after mutations happen, and then they get rid of all the other copies they don’t need. And then it collapses again,” he said.
“In the long term, you find these extremes. You find a pandoravirus, for example, which has apparently had tremendous genome expansion, and it’s 2.5 megabases, all with genes that we don’t really recognize because they are mostly only found in that lineage of viruses. So, for example, by gene duplication and diversification, you don’t recognize the genes anymore or where they came from. And this is specific for each subgroup of giant viruses.”
Genomic arsenals
Why these viruses need such massive genomes, however, remains unclear.
“Maybe that enables them to quickly switch hosts and adapt to many different intracellular environments if they’re already bringing a large set of genes,” Fischer said. “Combine that with the genomic flexibility — that they can lose genes and take off genes from the hosts — that makes them very versatile and adaptable organisms, right?”
Whether those viruses are able to utilize their genome fully is another question — one Steven Wilhelm, an environmental microbiologist at the University of Tennessee, Knoxville, recently set out to answer. In research published in Frontiers in Microbiology, he and his colleagues performed a transcriptomic analysis of the infection of the bloom-causing algae Aureococcus anophagefferens with its giant predator virus, AaV.
“One obvious question is, Do the viruses actually use these genes? Are they just tagging along like spare baggage? Or are the viruses actually transcribing them?” Wilhelm said. “We detected all but, I believe, three of the genes in the entire virus genome, which was kind of surprising to me. I thought there would be genes that weren’t being used, but that wasn’t the case.”
And AaVs constantly are refreshing their genetic arsenal to keep up with mutations, born out of natural selection, that allow surviving Aureococcus anophogefferens to subvert them. Wilhelm likens this genomic arms race to the Red Queen in Lewis Carroll’s “Through the Looking-Glass.”
“She stopped at one point and quipped, ‘Sometimes it takes all the running you can do just to stay in the same place.’ And we think about these virus host relationships like that,” Wilhelm said. “The viruses are trying to evolve constantly to affect all the hosts, and hosts are being pushed away from the viruses by selection and evolving. And the two of them keep moving forward to just stay in one place.”
Viral bloom

By numbers alone, viruses rule the oceans. They also outnumber every other microorganism in lakes, rivers and rivulets.
“In every environment, you find somewhere around 10 million virus particles per milliliter (of water),” Fischer said. “It doesn’t matter whether you go out in the open ocean, whether you go to the bottom of the ocean, whether you go to freshwater ponds, soil samples — they pretty much always have viruses as the most abundant biological entities.”
“The big ecological question is, What is the effect of these viruses, right? Now, do they all kill the hosts and are bad for them?” Fischer said. “No, that doesn’t seem to be the case. Instead, it looks like they’re fueling the ecosystem by providing nutrients.”
Giant marine viruses such as Wilhelm’s quarry AaV accomplish this by breaking up blooms of phytoplankton, whose bodies then rain down to the seafloor to sequester carbon. By doing so, AaVs return valuable phosphorus, iron and nitrogen that had been hoarded by a handful of highly competitive bacteria in the food web.
“Without viruses, all this would stagnate, basically. And you would get a few winning microbes that just do the same thing,” Fischer said. “And also, the diversity would be much lower, because viruses affect the most abundant hosts.”
Last year, two Tupanviruses, named after Tupã, the indigenous South American Guarani god of thunder, that had been isolated from water samples off the coast of Brazil and at the bottom of an alkaline lake bed in the same country, were described by a team of Brazilian and French scientists in a paper in Nature Communications. The viruses, members of the viral family Mimiviridae, possess the long tails, or fibers, that confounded Xiao in his days at Purdue as well as extraordinarily complex translational apparatuses.
While dozens of giant viruses are discovered each year, far fewer are characterized and added to the tree of terrestrial ancestry. Last year, researchers at the U.S. Department of Energy’s Joint Genome Institute brought mini-metagenomics tools to bear on soil samples from Harvard Forest, a 4,000-acre ecological research area in Petersham, Massachusetts, discovering 16 novel giant viruses.
Wilhelm chalks the boom up to technological growth.
“That, in part, is due to the democratization of tools for high-throughput molecular biology and bioinformatics and our ability to understand how to use these tools better,” he said.
Unfortunately, the time it takes for each newly identified giant virus to be characterized — its membrane, its genome, its relatives ancient and extant — is not insignificant, especially for a field with fewer than a dozen senior researchers.
La Scola, the virus hunter in Marseille who was the first author on the 2003 paper that identified Mimivirus as a virus and the senior author on the paper identifying Tupanvirus, has a backlog that could take him more than two decades to work through, collaboration and technology notwithstanding.
“In my freezer, I have 24, 25 completely new giant viruses, but I have no time to publish about them,” La Scola said. “Each time you find a virus, to describe it is probably one year of work, between microscopy and all the genome analysis.
Continuing resolution
At UTEP, Xiao continues to examine the mechanisms CroV uses to assemble its capsid. Though he has ample access to cryo-EM facilities, he worries about running up against his computer’s memory restrictions while reconstructing the virus’ capsid.
“I don’t know how long it would take us to finish the reconstruction, because we immediately hit a memory issue,” he said. “One terabyte of memory, which is the biggest they can put on the current supercomputer, isn’t enough. If we just look at the protein shell, if we count all of the hydrogens, it’s about 120 million atoms. A couple of years ago, when I tried to load and visualize 120 million atoms, my computer crashed.”
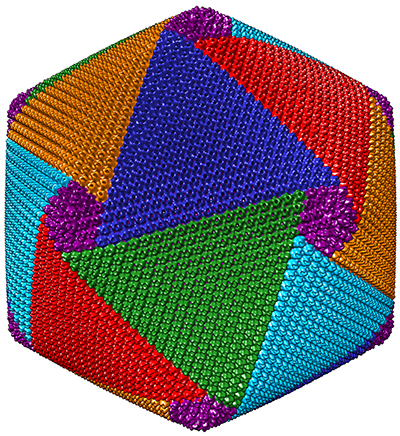
About two years ago, Xiao and his colleagues discovered that CroV assembles its protein shell in a spiral pattern made up of five interlocking vertices. They hope eventually to develop therapeutic viruslike nanoparticles by refining their resolution of CroV’s capsids.
“That’s what my new project is really focused on,” Xiao said. “We need to push the resolution higher.”
At Purdue, Klose and Richard J. Kuhn are carrying the torch from the lab of Xiao’s former PI, Michael Rossman, and continuing to bring the cryo-EM facility’s modern, fully digital electron microscopes to bear on pathogenic viruses, including Zika, West Nile and hepatitis C.
But their work puzzling out understudied viruses still is informed by and dependent on the discoveries of their colleagues.
“It’s kind of challenging for a lot of these projects to establish the right foundation, because we do structural biology so we don’t do any of the proteomics or genomics part, and we rely on our collaborators,” Klose said. “But if they’ve moved on to the next giant virus, it can be sometimes very difficult for us to get the information that we need.”
Basic research can be a hard sell when it comes to pulling in grants from the National Institutes of Health and National Science Foundation, something Klose predicts may become more difficult without Rossmann. While interrogating the capsids of giant viruses sometimes provides insight into structures with relevance to pathogenic viruses — as with CroV and influenza viruses, or with African swine fever and faustovirus, an NCLDV that Klose and Rossmann reconstructed a few years ago — those connections can’t be predicted.
“In the last few years, we have made a lot of progress in understanding what actually holds these (giant viruses) together and how they assemble,” Klose said. “But we also still have a long way to go to fully understand what’s going on and understand what these viruses do in the environment, which was always a question that Michael had.
“There are so many of them. They’re so diverse, they’re so big. They’ve been ignored for so many years. But what do they actually do?”
Viruses of viruses
Even viruses have predators. Over eons of acquiring and shedding genes, a number of giant viruses picked up smaller viruses known as virophages, whose DNA has infiltrated their own and springs to pseudo-life when the host virus begins its own transcriptional hijacking of bacteria.
“Complexity comes at a cost, right?” said Matthias Fischer, a microbiologist at the University of Heidelberg. “These giant viruses have acquired so many genes, and they can do so many things that other viruses rely on the host cell for, that now they’re paying the price for that by attracting this other type of parasite.
“One was isolated from a patient with keratitis — there was an amoeba causing an eye infection, the amoeba was infected with a giant virus, and that giant virus was infected with a virus.”
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
The science of staying strong
Muscles power every movement, but they also tell the story of aging itself. Scientists are uncovering how strength fades, why some species resist it and what lifestyle and molecular clues could help preserve muscle health for life.
Bacteriophage protein could make queso fresco safer
Researchers characterized the structure and function of PlyP100, a bacteriophage protein that shows promise as a food-safe antimicrobial for preventing Listeria monocytogenes growth in fresh cheeses.

Building the blueprint to block HIV
Wesley Sundquist will present his work on the HIV capsid and revolutionary drug, Lenacapavir, at the ASBMB Annual Meeting, March 7–10, in Maryland.
Gut microbes hijack cancer pathway in high-fat diets
Researchers at the Feinstein Institutes for Medical Research found that a high-fat diet increases ammonia-producing bacteria in the gut microbiome of mice, which in turn disrupts TGF-β signaling and promotes colorectal cancer.

Mapping fentanyl’s cellular footprint
Using a new imaging method, researchers at State University of New York at Buffalo traced fentanyl’s effects inside brain immune cells, revealing how the drug alters lipid droplets, pointing to new paths for addiction diagnostics.

Designing life’s building blocks with AI
Tanja Kortemme, a professor at the University of California, San Francisco, will discuss her research using computational biology to engineer proteins at the 2026 ASBMB Annual Meeting.

