Masters of physiology
This is an excerpt from Wilcox’s book “Venomous: How deadliest creatures mastered biochemistry,” which was published earlier this year by Scientific American/Farrar, Straus and Giroux. The chapter “Masters of physiology” opens with a description of the peculiarities of the platypus, one of the 12 venomous mammals, and then delves into the characteristics of venoms and why venomous animals invest the effort required to make them. The excerpt below, which has been edited for length and style, is the latter half of the chapter and describes how methods for venom analysis have changed over time.
 “Fight of the Mongoose and the Serpent Armies”, a watercolor depiction of the age-old battle between snake-eaters and their prey.IMAGE COURTESY OF ART RESOURCE
“Fight of the Mongoose and the Serpent Armies”, a watercolor depiction of the age-old battle between snake-eaters and their prey.IMAGE COURTESY OF ART RESOURCE
One of the problems with studying venoms until very recently was that we didn’t have very good ways of teasing apart what was present in the crude substances milked from animals. Chemists had nearly mastered the art of separating vastly different types of compounds, such as lipids and proteins, but such methods didn’t finely separate venom components. It was like sorting laundry: They could pull shirts from socks but couldn’t separate based on fabric color or distinguish long sleeves from short ones. Some venoms have hundreds of different peptides (small proteins), all of which might be soluble in water, for example. That means that an “aqueous fraction,” or subsection of a venom separated using water, might contain hundreds of different venom compounds, making it impossible to determine if one or many are responsible for any activity seen when that fraction is injected into a mouse.
 The speckled rattlesnake has impressive camouflage.PHOTO BY CHIP COCHRAN
The speckled rattlesnake has impressive camouflage.PHOTO BY CHIP COCHRAN
Luckily, back in the early 20th century, the Russian scientist Mikhail Tsvet invented a method to separate pigments from plants that came to be known as chromatography, which, with many later variations and refinements, has helped current scientists to isolate and identify venom components. In chromatography, mixtures are dissolved in a fluid (referred to as the mobile phase), which is then passed through a structure (the stationary phase) with certain properties. This structure can be simply a column of material through which the solution passes, drawn by gravity, or it can have specific chemical properties that make it “sticky” to particular types of molecules. When the mixture is run through the stationary phase, even small variations in the compounds’ size, 3-D structure or chemical properties cause molecules to travel at different speeds, allowing scientists to separate venom components on a much finer scale.
Throughout the 1940s and 1950s, new kinds of chromatography were invented, and what is now known as high performance liquid chromatography entered the scene. HPLC, which uses high air pressure instead of gravity to move the solution through a finer-textured column, is now one of the most important techniques in the study of venoms, as it allows scientists to separate venom samples into individual components. And conveniently, during the mid- 20th century, scientists also invented gel electrophoresis for separating molecules of protein, DNA or RNA. Gel electrophoresis uses an electric field to pull compounds through a gel by attracting negatively charged molecules to one end, while the gel’s properties affect which things will move through it more easily, traveling farther in a given amount of time. You can imagine how much faster a needle can be pushed into molasses than a finger can, for example, if they were both pressed with the same amount of force. When it comes to proteins, electrophoresis is mostly used to separate based on size, giving scientists a rough idea of the number of different proteins present in a venom. It also has become an invaluable method for determining whether genetic extractions or amplifications were successful, and is an absolute requirement in just about every lab that studies venom today.
 Komodo dragons are a species of venomous lizards.PHOTO BY CHRISTIE WILCOX
Komodo dragons are a species of venomous lizards.PHOTO BY CHRISTIE WILCOX
The modern era of venom research followed on the heels of these two major advances in separation technology. By the 1970s, labs worldwide could examine different components of a venom and their individual activities rather than the crude venom as a whole, and they began separating out the ones responsible for the most noticeable venom actions. Captopril—one of the best-selling drugs of all time, used to treat high blood pressure and heart failure—was isolated (from the venom of a Brazilian pit viper, Bothrops jararaca) during this time, as were many other venom compounds.
As a part of his Ph.D. thesis published in 1973, Peter Temple-Smith took advantage of the new battery of techniques to determine the contents and activities in platypus venom. He found at least 10 different proteins through electrophoresis and chromatography, and isolated the components that were lethal in mice from ones that caused convulsions. However, the scope of his research was limited, as the separation methods and bioassays still required relatively large amounts of venom (Temple-Smith couldn’t complete lethality tests, for example, because he didn’t have enough venom to work with). Snakes are easy, as they can be milked repeatedly and produce milliliters, and even liters, of venom fairly readily, but many of the other groups of venomous animals provide only 1/1000th or less of the volume required to run such tests. Though the platypus is capable of delivering upwards of 4 milliliters of venom with each spur, actually getting that much raw material is exceptionally difficult. On average, Temple-Smith and others found they could extract only 100 microliters at a time—too little, in those days, for detailed analyses.
But soon enough, tests miniaturized, and better technologies emerged to determine the shape and structure of different molecules, removing the large-volume requirements that had hindered progress. Scientists who made advances in mass spectrometry and nuclear magnetic resonance won Nobel Prizes in chemistry, and for the first time, these advances allowed them to deduce the chemical composition of larger, more complex compounds like those found in venoms. Even small volumes of crude venom could be evaluated to find compounds that are responsible for key activities such as reducing blood pressure, shutting down nerve impulses, or destroying red blood cells.
In the 1990s, several studies picked up where Temple-Smith left off. Scientists taking a closer look at platypus venom isolated active peptides, two different proteases and a hyaluronidase (enzymes also referred to as venom “spreading factors” because they cut hyaluronic acid, a major component of skin and the connective “goo” between cells). They could even obtain short sequences from some of these components, and determine that they are similar to snake venom constituents.
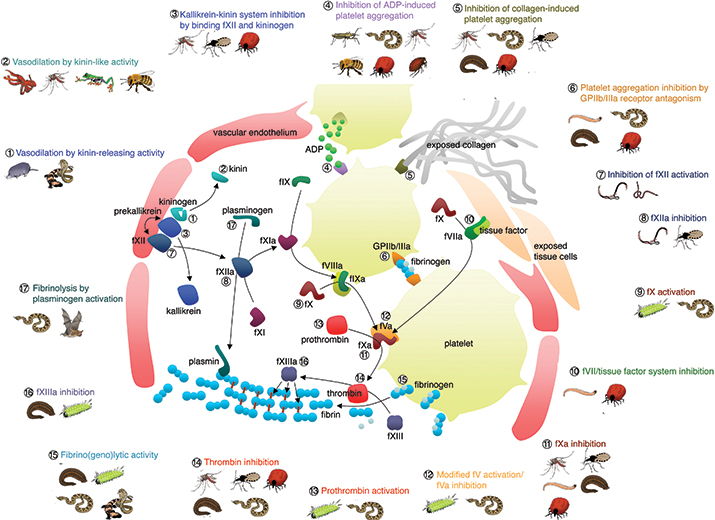 The diversity of hemotoxic venom compounds which disrupt the coagulation cascade.FIGURE BY BRYAN GRIEG FRY
The diversity of hemotoxic venom compounds which disrupt the coagulation cascade.FIGURE BY BRYAN GRIEG FRY
Then a new technology completely changed the way in which scientists study venomous animals and their toxins: genomics. (James) Watson, (Francis) Crick, and (Rosalind) Franklin had deduced the structure of DNA in 1953. The first sequencing technology, Sanger sequencing, came about in the 1970s followed by a method for amplifying fragments of DNA based on their sequence. The first full gene was sequenced in 1989, and the first full non-viral genome (a bacterium) in 1995. In the 20 or so years since, genetics and genomics have proved to be among the most rapidly changing fields in science. High-throughput technologies can now sequence entire genomes in a matter of hours, and new methods are regularly introduced that produce more information in less time for a lower price. It took years and cost millions to sequence the first human genome, which was finished in 2003—and it’s possible that within the next five to 10 years, sequencing an entire human genome will cost less than $1,000.
When it came to studying venoms, the genetics revolution opened up avenues that had never been imagined. Scientists could use genes to look at evolutionary relationships, and determine which species were closely related. They could compare the sequences of toxins to other proteins, and begin to understand how venoms evolve. And it wasn’t just DNA—scientists have developed methods of sequencing ribonucleic acid, the step in between DNA and proteins, and can determine which genes are being expressed. Genomics meant that they could sequence every protein expressed in a venom gland to look at the composition of a venom even without a single drop of it. Drug companies can build libraries of venom toxins and search them for ones that might act as enzymes, or have the potential to interact with a “target” such as an ion channel. By combining venom separation and component isolation with genomics, researchers have shifted from the study of venoms to venomics. Through such integrated research, we have come to know venomous animals far more intimately than at any point in history, and we have learned that their biochemical prowess is far more impressive than we ever imagined.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Using DNA barcodes to capture local biodiversity
Undergraduate at the University of California, Santa Barbara, leads citizen science initiative to engage the public in DNA barcoding to catalog local biodiversity, fostering community involvement in science.

Targeting Toxoplasma parasites and their protein accomplices
Researchers identify that a Toxoplasma gondii enzyme drives parasite's survival. Read more about this recent study from the Journal of Lipid Research.

Scavenger protein receptor aids the transport of lipoproteins
Scientists elucidated how two major splice variants of scavenger receptors affect cellular localization in endothelial cells. Read more about this recent study from the Journal of Lipid Research.

Fat cells are a culprit in osteoporosis
Scientists reveal that lipid transfer from bone marrow adipocytes to osteoblasts impairs bone formation by downregulating osteogenic proteins and inducing ferroptosis. Read more about this recent study from the Journal of Lipid Research.

Unraveling oncogenesis: What makes cancer tick?
Learn about the ASBMB 2025 symposium on oncogenic hubs: chromatin regulatory and transcriptional complexes in cancer.

Exploring lipid metabolism: A journey through time and innovation
Recent lipid metabolism research has unveiled critical insights into lipid–protein interactions, offering potential therapeutic targets for metabolic and neurodegenerative diseases. Check out the latest in lipid science at the ASBMB annual meeting.

