MCP: Guidelines for reporting
complex spectra
The editors of the journal Molecular & Cellular Proteomics are seeking feedback from the proteomics community on draft guidelines for publishing proteomics studies that use data-independent acquisition, or DIA, methods. The draft guidelines can be accessed from the journal’s homepage. Comments will be accepted until the end of September.
Steven Carr is deputy editor of MCP and senior director of proteomics at the Broad Institute. “DIA is a rapidly growing research approach that can be employed on a wide variety of instrument platforms,” he said. “As such, it is important to establish rules to make sure it is properly applied.”
DIA is used to collect tandem mass spectrometry data. It offers broad coverage of the proteome with high run-to-run reproducibility.

Mass spectrometers sort the ions from a sample based on their mass-to-charge ratio. In tandem mass spectrometry, there are two ionization and sorting steps. For reliable identification of a molecule, you need to know its intact precursor ion mass and also the masses produced when it is fragmented. Most strategies involve fragmenting a single precursor ion at a time.
Selection of a precursor ion can be based on observation of a peak in the first spectrum (a strategy known as data-dependent acquisition, or DDA), or it can be from a list of predetermined components of interest, i.e., targeted analysis. However, both of these approaches select only a subset of the components present for fragmentation analysis. In DIA, the whole mass range is fragmented over a series of scans. This provides fragmentation information about all components, but fragmenting multiple components at the same time produces complex spectra made of heterogeneous precursors.
With technical advances in instrumentation and computation, DIA approaches are growing in popularity, particularly for quantitative studies of sets of related samples. However, because the spectra collected using DIA are significantly more complex than data from other approaches, they can be more difficult to interpret. Complicating the situation further, there are many competing techniques for collecting DIA data, few of which have been compared directly, and researchers have yet to develop fieldwide standards around how to interpret and report results.
MCP is taking steps to ensure that future data will be described more systematically. The editors brought together 25 DIA experts from academia and industry at a satellite workshop in San Diego after the close of the American Society for Mass Spectrometry’s 2018 meeting in June. The guidelines drafted at this workshop aim to help researchers write a thorough description of how DIA data were collected and interpreted, rendering researchers’ conclusions easier to evaluate.
Among the workshop’s organizers was Robert Chalkley of the University of California, San Francisco, MCP data management editor. “When we publish the draft, we will give the opportunity for anyone in the community to send in their comments and suggestions,” he said.
MCP’s reporting guidelines for other types of mass spectrometry study have been adopted widely by other journals. MCP’s editors hope to continue to lead the field in producing guidelines that aim to allow independent assessment of the reliability of published data sets.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.
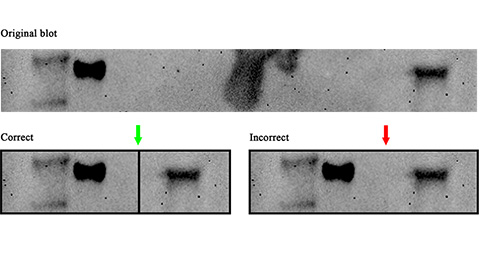
Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.
Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.
Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.
Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

