New studies define transcription–translation coupling
Given the crowd of macromolecules squeezed higgledy-piggledy into cells, it’s sometimes surprising that any multi-step pathway reaches completion. But coordination emerges from the soup, and sometimes many pathways work in tandem.
Three structural studies of coupling between transcription and translation in bacteria published in the journal Science — two of which came out today — are rearranging how both RNA polymerase and ribosome experts think about their fields. The work also shows off the dazzling power of cryo-electron microscopy.
Jamie Cate, a ribosome biologist at the University of California, Berkeley expects these papers to change the way both complexes are studied. “One of the things about this series of papers is that it’s bringing together fields that have been able to live happily separate lives … but now it’s clear that they can’t do that anymore; they’ve got to come together,” Cate said.
To copy genetic code into a translatable format, RNA polymerase, or RNAP, skates along a bubble in the DNA, busily matching new RNA bases to the DNA template to build the strand of messenger RNA. In bacteria, which lack the orderly formality of a nucleus, the much-larger ribosome bumps along at RNAP’s heels, translating messenger RNA into a chain of amino acids that will fold into functional proteins.
Some DNA sequences cause RNAP to stall out, and some mRNA sequences trip up the ribosome. Researchers have observed that when the two complexes are together, both are more successful in moving forward. And as early as the 1970s, electron microscopists noticed that ribosomes and polymerases could be found in close proximity, as what one researcher called “two blobs … that look really close.”
Because both are made of many moving parts, structural biologists have had difficulty getting a closer look at the two blobs and understanding how they fit together. The recent spate of studies offers insight into how a comparatively tiny protein hitches the behemoths together and what the coupling of processes might mean for bacterial biology.
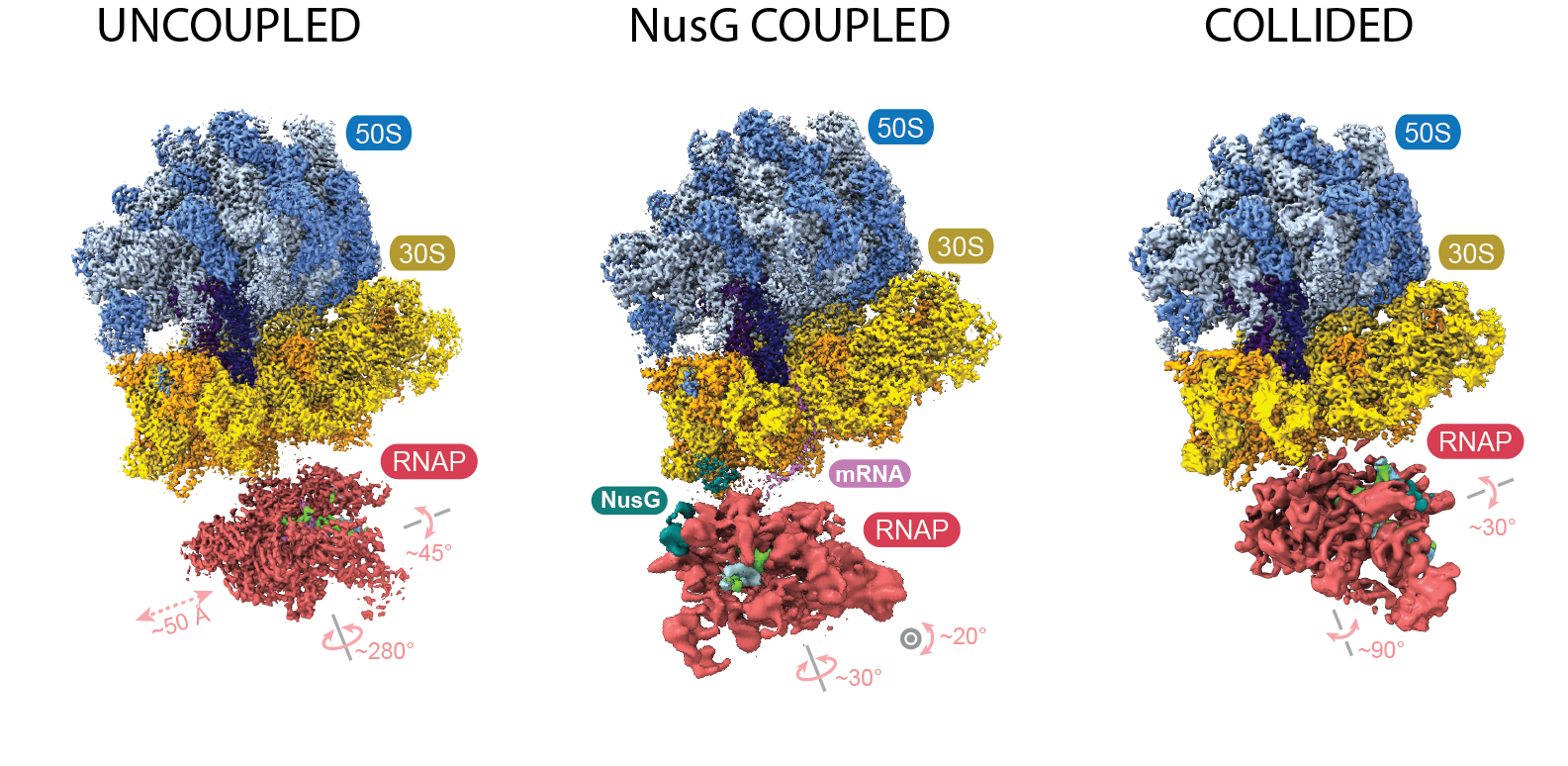
Digging for diamonds
Structural biologists in Patrick Cramer’s and collaborator Bob Landick’s labs were among the first to find a way to use high-resolution microscopy techniques to study the transcription–translation complex. In a study published in 2017, they slammed the brakes on an RNA polymerase in a test tube, letting the trailing ribosome crash into it. Using cryo-electron microscopy — a technique that depends on flash-freezing a solution, capturing electron micrographs of its jumble of particles, and using the collected images to reconstruct how those particles look in 3D — they obtained the first high-resolution structure of the E. coli ribosome–polymerase complex.
“Like all good results do,” Landick said, the structure left them with “more questions than there are answers.”
Years of biochemical and genetic work had suggested that a universally conserved protein called NusG, which is tiny compared to the great masses of ribosome and polymerase, forms a tether between the two. But in the 2017 structure, there was no place for that tether to fit. That prompted Rutgers professor Richard Ebright and his lab to look into why results from the two types of studies conflicted.
Instead of colliding the ribosome into the polymerase, Ebright’s team gave both complexes a well-defined mRNA scaffold, with just one place for each to bind. They expected that the length of that scaffold might determine whether there was space for NusG to fit.
At first, their structure, like the earlier one, showed ribosome and RNAP in direct contact, with no place for NusG. They repeatedly lengthened the scaffold, making complicated mixtures of more than a dozen purified components. After each cycle, which took about a month from recipe to final dataset, they found the same collided structure.
“This project seemed like mining,” first author Chengyuan Wang, a postdoc in Ebright’s lab, said. “You don’t know where the diamond is; you just dig, dig, dig.”
They decided to try one final scaffold, their longest yet, and that’s when they saw something different. NusG was included in this structure, poised like a towrope between RNAP and the ribosome. The RNA’s length was important after all.
A shoulder angel
When Wang and a colleague from Ebright’s lab presented their work at a meeting last year, they met Albert Weixlbaumer, a principal investigator at the French research institute IGBMC. They had no idea his lab was doing similar work.
“I had a devil on one shoulder, an angel on the other shoulder,” Weixlbaumer said. “The devil told me, ‘Don’t tell them anything! Don’t tell them anything!’ and the angel said ‘No, the best thing to do is to coordinate.’”
By then, Weixlbaumer’s lab had been working for several years to obtain a cryo-EM structure of the transcription-translation complex. The team, like Ebright’s, observed what they called collided, coupled and uncoupled structures. The collided structure closely resembles the one published in 2017; the coupled structure shows the ribosome and RNAP in close proximity, but not touching, with NusG between them; and in the uncoupled structure, the two complexes make no contact at all.
The two groups coordinated on today’s publication of their findings in Science. The structures they observed, while very similar, still have some differences. Both showed that NusG is an important tether between the coupled polymerase and ribosome, but only Ebright’s team included a second cofactor, NusA, in the mix. They saw that it, too, fits into the coupled RNAP-ribosome structure, and propose that it might be important for maintaining appropriate distance between the complexes.
Both labs also captured lower-resolution images of many other structures that RNAP and the ribosome can adopt. They haven’t described them in detail, but can see that they exist — and the scientists are intrigued by the biochemical implications.
“There seem to be so many different ways in which the two complexes come together,” said Michael Webster, a postdoc in the Weixlbaumer lab. The crucial question going forward is whether and how each of these structures function in living cells.
Coupling in cells
Recently, European researchers managed to capture images and model a complex in the bacterium Mycoplasma pneumoniae that linked RNA polymerase with the ribosome — frozen in the act of transcribing and translating, inside a cell.
“It’s a technical breakthrough, no doubt about it,” Weixlbaumer said of the study. “It’s fantastic.”
Led by Julia Mahamid of EMBL–Heidelberg and Juri Rappsilber at the University of Technology, Berlin, this group used cryo-electron tomography, a technique related to cryo-EM, to collect 3D images of whole cells, integrated with crosslinking mass-spectrometry to identify the proteins they saw.
“In our case, the critical protein density between RNAP and the ribosome was not the protein everybody would have expected, NusG,” Rappsilber wrote in an email. Instead, they identified the tether as NusA.
The researchers looked at a simpler organism than E. coli; changes that make the mycoplasma easier to image, such as its stripped-down genome, may also make its protein expression machinery an imperfect match for those that most in vitro transcription–translation studies have used.
“This convergence of new structures is definitely taking the field to another level,” said Cate of the three articles. “But it’s far from the end of the story … because these test tube experiments and the cell-based experiments don’t fully line up.”
The molecular details may vary, colleagues say, but Rappsilber and Mahamid’s work illuminates fascinating biology. Having proved that transcription–translation coupling can occur in living cells, they also showed that an antibiotic that inhibits translation can disrupt formation of the coupled complex — suggesting that the drug may affect transcription too, and potentially altering our understanding of many drugs’ mechanisms of action.
Other scientists are most excited about the possibilities for using advanced imaging techniques to investigate molecular machines in their real, messy environments.
“I think we’ve just begun to scratch the surface of what can be done,” Landick said.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Using DNA barcodes to capture local biodiversity
Undergraduate at the University of California, Santa Barbara, leads citizen science initiative to engage the public in DNA barcoding to catalog local biodiversity, fostering community involvement in science.

Targeting Toxoplasma parasites and their protein accomplices
Researchers identify that a Toxoplasma gondii enzyme drives parasite's survival. Read more about this recent study from the Journal of Lipid Research.

Scavenger protein receptor aids the transport of lipoproteins
Scientists elucidated how two major splice variants of scavenger receptors affect cellular localization in endothelial cells. Read more about this recent study from the Journal of Lipid Research.

Fat cells are a culprit in osteoporosis
Scientists reveal that lipid transfer from bone marrow adipocytes to osteoblasts impairs bone formation by downregulating osteogenic proteins and inducing ferroptosis. Read more about this recent study from the Journal of Lipid Research.

Unraveling oncogenesis: What makes cancer tick?
Learn about the ASBMB 2025 symposium on oncogenic hubs: chromatin regulatory and transcriptional complexes in cancer.

Exploring lipid metabolism: A journey through time and innovation
Recent lipid metabolism research has unveiled critical insights into lipid–protein interactions, offering potential therapeutic targets for metabolic and neurodegenerative diseases. Check out the latest in lipid science at the ASBMB annual meeting.

