JBC: How bacteria build efficient photosynthesis machines
Researchers facing a future world with a larger human population and more uncertain climate are looking to photosynthetic bacteria for engineering solutions to improve crop yields.
In the Journal of Biological Chemistry, a Canadian research team reports on how cyanobacteria finesse one of the most wasteful steps in photosynthesis. The study investigated the assembly of carboxysomes in which the bacteria concentrate carbon dioxide, boosting the efficiency of a critical enzyme called RuBisCO.
“Essentially everything we eat starts with RuBisCO,” said Matthew Kimber, a professor at the University of Guelph in Ontario and senior author on the paper.
The enzyme, which is made of 16 protein subunits, is essential for photosynthesis. Using energy captured from light, it incorporates carbon dioxide into organic molecules from which a plant then builds new sugar. Unfortunately, it’s not terribly efficient. Or, from Kimber’s point of view, “RuBisCO has a really thankless task.”
The enzyme evolved in an ancient world where carbon dioxide was common and oxygen was rare. As a result, it isn’t very picky in discriminating between the two gases. Now that the atmospheric tables have turned, RuBisCO often accidentally captures oxygen, generating a useless compound that the plant then has to recycle.
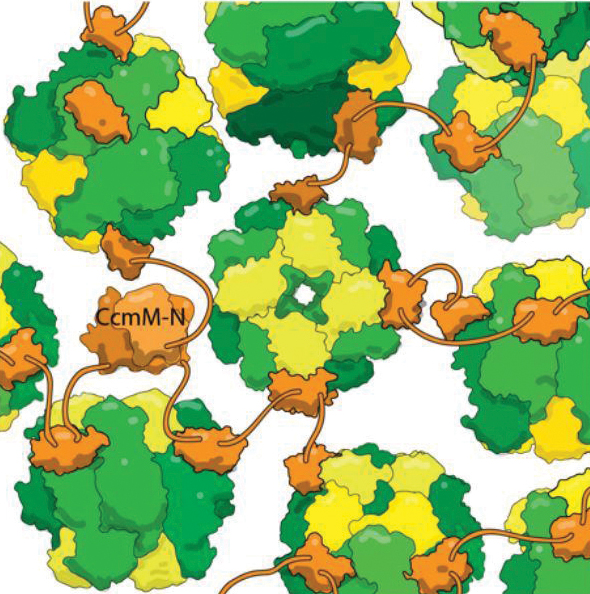
Cyanobacteria make few such mistakes, because bacteria collect their RuBisCO into dense bodies known as carboxysomes. The bacteria pump bicarbonate (simply hydrated CO2) into the cell; once it gets into the carboxysome, enzymes convert the bicarbonate into carbon dioxide. Because the carbon dioxide can’t escape through the protein shell surrounding the carboxysome, it builds up to high concentrations, helping RuBisCO avoid costly mistakes.
Kimber wants to understand the logic of carboxysomes’ organization. “They’re actually phenomenally intricate machines,” he said. “The cyanobacterium makes 11 or so normal-looking proteins, and these somehow organize themselves into this self-regulating mega-complex that can exceed the size of a small cell.”
One of carboxysomes’ most impressive tricks is self-assembly, which Kimber’s lab set out to understand. They looked at a protein, CcmM, that corrals RuBisCO enzymes into new carboxysomes. They knew that part of CcmM looks a lot like a subunit of RuBisCO — so much so that researchers suspect ancient cyanobacteria created CcmM by duplicating a RuBisCO gene.
Most scientists in the field believed that CcmM binds to the enzyme by usurping that RuBisCO subunit’s spot. But when Kimber’s lab took a detailed look at CcmM’s structure and binding, the results showed that was wrong. True, CcmM was similar in shape to the small RuBisCO subunit. But the complexes it formed still included all eight small subunits, meaning that instead of stealing a spot from a RuBisCO subunit, CcmM had to be binding elsewhere.
“This is very odd from a biological perspective, because if CcmM arose by duplicating the small subunit, it almost certainly originally bound in the same way,” Kimber said. “At some point, it must have evolved to prefer a new binding site.”
The researchers also found that a linker between binding domains in CcmM is short enough that “instead of wrapping around RuBisCO, it tethers (individual enzymes) together like beads on a string,” Kimber said. “With several such linkers binding each RuBisCO at random, it crosslinks everything into this big glob; you wrap a shell around it, and this then becomes the carboxysome.”
Scientists at another university reported last fall that they had succeeded in making tobacco plants with a stripped-down carboxysome in their chloroplasts. Those plants didn’t grow especially well, and the authors concluded that they had taken away too many components of the carboxysome; although it could be built in the chloroplast, it was a drag on the plants instead of a help.
A better understanding of how proteins like CcmM contribute to carboxysome construction and function could help bioengineers leverage carboxysome efficiency in the next generation of engineered plants.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
Cracking cancer’s code through functional connections
A machine learning–derived protein cofunction network is transforming how scientists understand and uncover relationships between proteins in cancer.

Gaze into the proteomics crystal ball
The 15th International Symposium on Proteomics in the Life Sciences symposium will be held August 17–21 in Cambridge, Massachusetts.
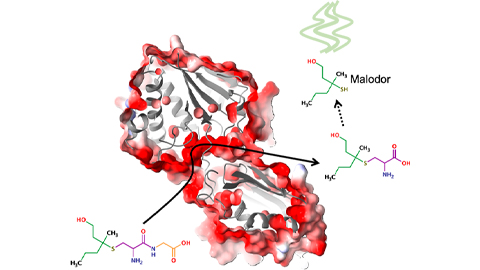
Bacterial enzyme catalyzes body odor compound formation
Researchers identify a skin-resident Staphylococcus hominis dipeptidase involved in creating sulfur-containing secretions. Read more about this recent Journal of Biological Chemistry paper.

Neurobiology of stress and substance use
MOSAIC scholar and proud Latino, Bryan Cruz of Scripps Research Institute studies the neurochemical origins of PTSD-related alcohol use using a multidisciplinary approach.
Pesticide disrupts neuronal potentiation
New research reveals how deltamethrin may disrupt brain development by altering the protein cargo of brain-derived extracellular vesicles. Read more about this recent Molecular & Cellular Proteomics article.

A look into the rice glycoproteome
Researchers mapped posttranslational modifications in Oryza sativa, revealing hundreds of alterations tied to key plant processes. Read more about this recent Molecular & Cellular Proteomics paper.

