‘Filling the void of the virosphere’
Just one RNA virus, SARS-CoV-2, has changed the lives of billions of people in the past three years. What if I told you that a recent expedition found more than 5,500 new species of RNA viruses in oceans across the globe? Don’t be nervous. There’s little for humans to worry about. RNA viruses in the ocean are doing a lot more than just killing their hosts; they have a role in maintaining the equilibrium of the ocean ecosystem and may even help mitigate climate change.
The discovery of the new viruses was reported in the journal Science in April. The research team, led by microbiology professor Matthew Sullivan at Ohio State University, analyzed samples collected a decade ago by the Tara Oceans expedition.
Combining machine learning approaches, the team built a phylogenetic tree that, in the end, doubled the number of phyla (groups of species with similar characteristics) of RNA viruses from five to 10.
Guillermo Dominguez–Huerta, a postdoctoral scholar in Sullivan’s lab and a co-author on the paper, said, “What excites me the most is that we’re closer to understanding the real history of evolution and ecology of RNA viruses — what is happening with viruses in nature, specifically in the oceans.”

Tara: The unsung hero
The 36-meter schooner was built in 1989 for French doctor and explorer Jean-Louis Étienne, who just a few years earlier had become the first person to reach the North Pole by skiing solo. He used the ship to study the polar regions in Antarctica, which is why its original name was Antarctica.
A decade later, it was purchased by a famous regatta sailor, Peter Blake, who named it Seamaster and went on to win the America’s Cup, the oldest international competition in sailing, twice for his native New Zealand. After retiring from sailing and driven by his love of oceans, Blake used the Seamaster to advocate for the importance of the oceans until he was fatally shot by pirates off the Brazilian coast in 2001.
French fashion designer agnès b. and her son Étienne Bourgois acquired the ship, which they renamed Tara, in 2003 and founded the Tara Ocean Foundation to conduct scientific research for the protection of the ocean.

Tara left its French home port of Lorient for its first expedition in 2006 as part of the fourth International Polar Year, an interdisciplinary, collaborative and international program focused on research in the polar regions. With 11 crew members aboard collecting climate data, Tara spent months drifting with the sea ice of the Arctic Ocean.
“The Arctic Drift was a very difficult project. There were scientists on board who were alone for nine months,” said Chris Bowler, research director at the Institut de Biologie de l’École Normale Supérieure in Paris and chair of the scientific committee at the Tara Ocean Foundation. “But they all made it, and it was a huge success as a research program, setting the foundation for using the ship to explore marine biodiversity and its relation to climate.”
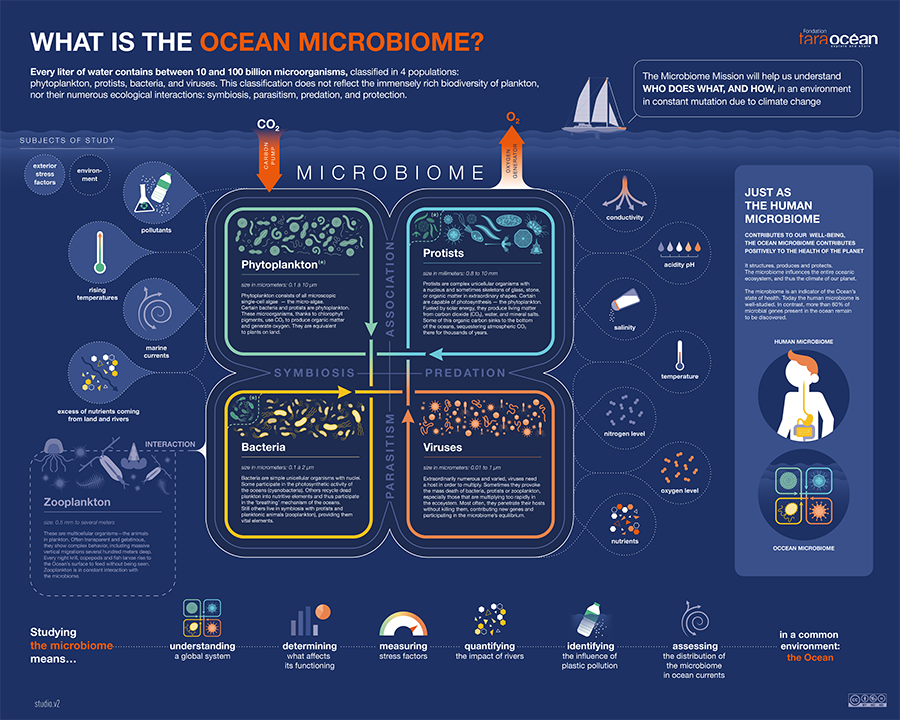
From 2009 to 2013, researchers aboard Tara collected samples from 210 sites in the Atlantic, Pacific, Arctic and Indian Oceans to showcase invisible aquatic life. At each sampling site, researchers sampled the whole water column from the surface to 1,000 meters below for all different kinds of microscopic life — from the smallest viruses (20 nm) to zooplankton (1-2 mm).
“These organisms cover five orders of magnitude. It’s like going from an ant to a brontosaurus in a forest ecosystem,” Bowler said. “These samples had been collected in a very standardized way, allowing ocean researchers across the globe to compare everything with everything else, and that was the beauty of our sampling.”

Developing tools to study RNA viruses
Back in Ohio, Sullivan’s group had studied DNA viruses in the ocean and their role in nutrient cycling for decades. However, studies of RNA viruses in the ocean have lagged, primarily due to lack of tools to identify them with high confidence. Also, the fact that RNA is less stable than DNA in the environment didn’t help.
Previous efforts to classify RNA viruses focused mostly on ones that cause livestock, plant or human illnesses. “But for viruses in the ocean, we didn’t have any information about hosts, and there were no viral particles to study,” Dominguez–Huerta said.

So the team decided the starting point was to sequence the RNA from organisms present in the ocean and then distinguish the RNA of the hosts from the RNA of the viruses.
“The journey from the expeditionary part of the project completed in 2013 by the Tara schooner to coming up with a catalog of RNA viruses from the ocean took us nearly 10 years,” Bowler said.
The first hurdle was to establish a protocol to sequence the RNA from samples from the ocean, as they often are contaminated and degraded. The French National Sequencing Center (Genoscope) did that. The second challenge was to develop methods for analysis of the sequencing data, and that’s where the Sullivan lab’s expertise in bioinformatics and oceanic viruses was put in action.
The analysis to distinguish viral RNA was based on the RNA sequence of a signature gene called RNA-dependent RNA polymerase, or RdRp, which is unique to RNA viruses.
“RdRp has evolved for billions of years in RNA viruses and, hence, is highly divergent in its sequence, making it difficult to align through traditional methods,” Dominguez–Huerta said. So the team resorted to using machine learning to organize the divergence of RdRp sequences by aligning only the functional domain of the protein, which should most accurately reflect its evolution.
In addition to the primary sequence of the RdRp, the team looked at the different kinds of genes the RNA viruses had, their genome architecture, and 3D structure of the RdRp to confirm the identification of five new phyla.
The most abundant newly identified species belong to a proposed phylum fittingly named Taraviricota.
“An intriguing feature of the phyla is that the 3D structure of the RdRp is very similar to reverse transcriptase (RNA-dependent DNA polymerase), suggesting that they might be a missing link in early RNA virus evolution and the origin of life,” Dominguez–Huerta said.
RNA viruses and climate change
What are these viruses doing in the ocean? In a follow-up study, the Ohio team determined that these viruses predominantly infect microbial eukaryotes, such as protist and fungal hosts, plus a few invertebrates.

These viral hosts play an important role in carbon export — the process by which carbon is pulled from the atmosphere, fixed into marine organisms and then exported to the depths of the ocean as those organisms sink to the seafloor. By infecting these organisms, RNA viruses likely affect how carbon flows through the ocean at large.
RNA viruses also may drive carbon flux by splitting their hosts open during lysis and spilling sequestered carbon into the ocean.
“The team also unexpectedly discovered that 95 of the RNA viruses carried genes they’d ‘stolen’ from their host cells,” Dominguez–Huerta said in an interview with Live Science.
In the host, these genes help to direct metabolic processes within the cell and hence were named auxiliary metabolic genes, or AMGs. This discovery suggests that the viruses manipulate their hosts’ metabolisms to maximize the production of new virus particles and evade host immune responses.
The future for Tara and oceanic viruses
What the scientists aboard Tara and in labs on land have achieved is incredible, but the sampling is a mere snapshot of ocean diversity.
“What we have to do is to go back and do more longitudinal time series — to understand how the ocean is changing and what the future of the ocean looks like,” Bowler said.
Dominguez–Huerta added: “This project was hard — to come up with the tools to identify viruses and make sure they’re validated — but we’re still very much at the beginning. We know about less than 1% of RNA viruses on the Earth. So many questions still need to be answered. Here, we’re providing a roadmap for other researchers to start filling the void of the virosphere.”

Reflection on the Tara Oceans expedition

Michael Sieracki of the National Science Foundation was among those aboard the Tara during the Tara Oceans expedition. In 2015, he published a reflection on the experience in the Bulletin of the Association for the Sciences of Limnology and Oceanography. Here’s an excerpt:
“In some ways, Tara Oceans resembled an oceanography of the past — a throwback to the heroic era of expeditionary science — like the Challenger expedition or the voyage of the Beagle, or the early mountaineers and polar explorers. These days we get these experiences vicariously through robots on Mars, or GoPro videos of extreme gravity sports. To young scientists coming into our field I must say that the adventure of oceanography is not over. We must stay open to new ways of envisioning it and open to the risks to make it happen. Discovery is the soul of science. The rewards could be better than you can imagine.”
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Targeting Toxoplasma parasites and their protein accomplices
Researchers identify that a Toxoplasma gondii enzyme drives parasite's survival. Read more about this recent study from the Journal of Lipid Research.

Scavenger protein receptor aids the transport of lipoproteins
Scientists elucidated how two major splice variants of scavenger receptors affect cellular localization in endothelial cells. Read more about this recent study from the Journal of Lipid Research.

Fat cells are a culprit in osteoporosis
Scientists reveal that lipid transfer from bone marrow adipocytes to osteoblasts impairs bone formation by downregulating osteogenic proteins and inducing ferroptosis. Read more about this recent study from the Journal of Lipid Research.

Unraveling oncogenesis: What makes cancer tick?
Learn about the ASBMB 2025 symposium on oncogenic hubs: chromatin regulatory and transcriptional complexes in cancer.

Exploring lipid metabolism: A journey through time and innovation
Recent lipid metabolism research has unveiled critical insights into lipid–protein interactions, offering potential therapeutic targets for metabolic and neurodegenerative diseases. Check out the latest in lipid science at the ASBMB annual meeting.
Melissa Moore to speak at ASBMB 2025
Richard Silverman and Melissa Moore are the featured speakers at the ASBMB annual meeting to be held April 12-15 in Chicago.