From the journals: MCP
An algorithm to treat leukemia, proteomic analysis of the substantia nigra and promiscuous binding in motor neuron disease. We offer summaries of papers on these topics recently published in the journal Molecular & Cellular Proteomics.
Algorithm identifies active kinases for AML treatment
Acute myeloid leukemia, or AML, is a cancer of the bone marrow and blood that progresses rapidly if left untreated. An estimated 19,000 new cases will be diagnosed this year with a five-year survival rate of 28.1%. Small-molecule kinase inhibitors are a potential treatment strategy for AML patients. However, due to molecular heterogeneity in AML, no single molecule has been clinically effective.
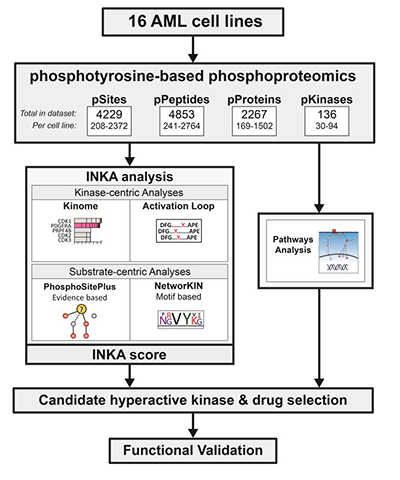
In their recent study published in the journal Molecular & Cellular Proteomics, Carolien van Alphen and colleagues at the Amsterdam University Medical Center relied on an integrative inferred kinase activity, or INKA, algorithm to identify prospective small-molecule kinase inhibitors. For each given kinase, INKA performs a four-component analysis by determining the phosphorylation status of the kinase itself, its activation loop and all its possible substrates. INKA assigns a nonzero score to kinases with both kinase-centric and substrate-centric phosphorylation status.
A panel of 16 AML cell lines was chosen for pY-phosphoproteomics combined with INKA analysis to identify hyperphosphorylated active kinases. Due to the heterogeneous signaling in these cell lines, the analysis identified multiple phosphopeptides. INKA analysis of individual cell lines identified specific driver kinases, including PDGFRA, JAK2, KIT and FLT3. Using this approach, the researchers identified and functionally verified active tyrosine kinases in 10 cell lines. For the remaining six cell lines without a tyrosine kinase driver, they identified MAPK signaling as a potential drug target.
The authors concluded the study by applying their analytical strategy to clinical samples. Despite the lower amount of input sample, phosphoproteomic and INKA analysis identified similar driver kinases in patient samples to those in the cell lines. The in-depth analysis performed in this study provides a basis for future clinical applications for personalized treatment of AML patients.
(Laser)-capturing the substantia nigra proteome
Many challenges hinder the analysis of tissue sections, including their heterogeneous nature and limited cell numbers in distinct cell populations. Laser capture microdissection, or LCM, is a technique that purifies small cell populations from mixed samples. In a study published in the journal Molecular & Cellular Proteomics, the authors present a detailed workflow for LCM-based proteomic analysis of the substantia nigra, a region of the brain rich in dopaminergic neurons and implicated in Parkinson’s disease. The authors have unraveled the substantia nigra proteome using limited quantities of tissue samples.Eva Griesser and colleagues at the University of Dundee, U.K., first optimized their protocol for isolating proteins from tissue samples. They performed comparative analysis for testing LCM efficiency in five healthy donors who each provided intact tissue and 3,000 cells, which were isolated with LCM. Bioinformatic analysis identified neuron-specific proteins and gene ontology, or GO, cellular components to be enriched in the microdissected samples.
Application of this optimized protocol to the substantia nigra from 15 healthy donors identified 5,677 protein groups with GO terms ranging from substantia nigra development and neurotransmitter secretion (high intensity) to lysosome and trans-Golgi network (low intensity). This methodology will facilitate proteomic analysis of different brain regions in the course of neurodegenerative disorders.
Promiscuous binding in motor neuron disease
Arginine-based dipeptide repeat polymers, or DPRs, are a unique characteristic of motor neuron disease, or MND. Among the abnormally expressed DPRs in patient brains, proline-arginine, or poly-PR, and glycine-arginine, or poly-GR, exhibit greater levels of cellular toxicity. The toxicity has been attributed to repeat-associated non-AUG-initiated translation that results in production of abnormal translation products of varying lengths. While it is reported that these polymers stall the ribosomal translation machinery and ribosome biogenesis, not much is known about the identity of the affected proteome. For a recent study published in the journal Molecular & Cellular Proteomics, Mona Radwan and colleagues at the University of Melbourne, Australia, performed whole proteomic profiling to probe the interacting partners of these toxic polymers of varying lengths.
Using a combination of immunoprecipitation, mass spectrometry and green fluorescent protein-labeled peptides, the authors identified multiple cellular pathways through which Arg-rich polymers manifest cellular toxicity. The affected protein machinery for poly-PR and poly-GR ranged from cytoskeletal proteins to arginine methylases PRMT1 and PRMT5, which was in striking contrast to the partners of inert polymers like poly-GA. The effect was further pronounced with varying lengths of these polymers, with 101x DPRs inducing marked ribosome stalling and toxicity. These insights into the multiple pathophysiological mechanisms inherent in MND pave a path for deeper understanding of neurodegeneration.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
The data that did not fit
Brent Stockwell’s perseverance and work on the small molecule erastin led to the identification of ferroptosis, a regulated form of cell death with implications for cancer, neurodegeneration and infection.

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.
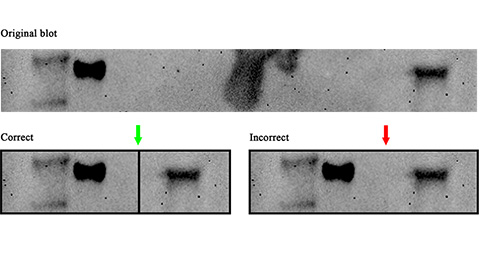
Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.
Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.
Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.
Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

