Revealing what makes bacteria life-threatening
Queensland researchers have discovered that a mutation allows some E. coli bacteria to cause severe disease in people while other bacteria are harmless, a finding that could help to combat antibiotic resistance.
Professor Mark Schembri and Dr Nhu Nguyen from IMB and Associate Professor Sumaira Hasnain from Mater Research found the mutation in the cellulose making machinery of E. coli bacteria. The research was published in Nature Communications.
Professor Schembri said the mutation gives the affected E. coli bacteria the green light to spread further into the body and infect more organs, such as the liver, spleen and brain.
"Bad' bacteria can't make cellulose
“Our discovery explains why some E. coli bacteria can cause life-threatening sepsis, neonatal meningitis and urinary tract infections (UTIs), while other E. coli bacteria can live in our bodies without causing harm,” Professor Schembri said.
“The ‘good’ bacteria make cellulose and ‘bad’ bacteria can’t.”
Bacteria produce many substances on their cell surfaces that can stimulate or dampen the immune system of the host.

Inflammation and spreading through the body
“The mutations we identified stopped the E. coli making the cell-surface carbohydrate cellulose and this led to increased inflammation in the intestinal tract of the host,” Professor Schembri said.
“The result was a breakdown of the intestinal barrier, so the bacteria could spread through the body.”
In models that replicate human disease, the team showed that the inability to produce cellulose made the bacteria more virulent, so it caused more severe disease, including infection of the brain in meningitis and the bladder in UTIs.
Finding new ways to prevent infection

Associate Professor Hasnain said understanding how bacteria spread from intestinal reservoirs to the rest of the body was important in preventing infections.
“Our finding helps explain why certain types of E. coli become more dangerous and provides an explanation for the emergence of different types of highly virulent and invasive bacteria,” she said.
Professor Schembri said E. coli was the most dominant pathogen associated with bacterial antibiotic resistance.
“In 2019 alone, almost 5 million deaths worldwide were associated with bacterial antibiotic resistance, with E. coli causing more than 800,000 of these deaths,” he said.
“As the threat of superbugs that are resistant to all available antibiotics increases worldwide, finding new ways to prevent this infection pathway is critical to reduce the number of human infections.”
This article was republished from the University of Queensland website. Read the original here.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

When oncogenes collide in brain development
Researchers at University Medical Center Hamburg, found that elevated oncoprotein levels within the Wnt pathway can disrupt the brain cell extracellular matrix, suggesting a new role for LIN28A in brain development.
The data that did not fit
Brent Stockwell’s perseverance and work on the small molecule erastin led to the identification of ferroptosis, a regulated form of cell death with implications for cancer, neurodegeneration and infection.

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.
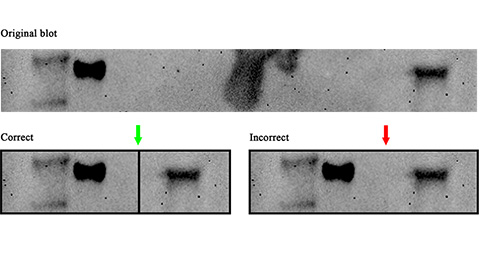
Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.
Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.
Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

