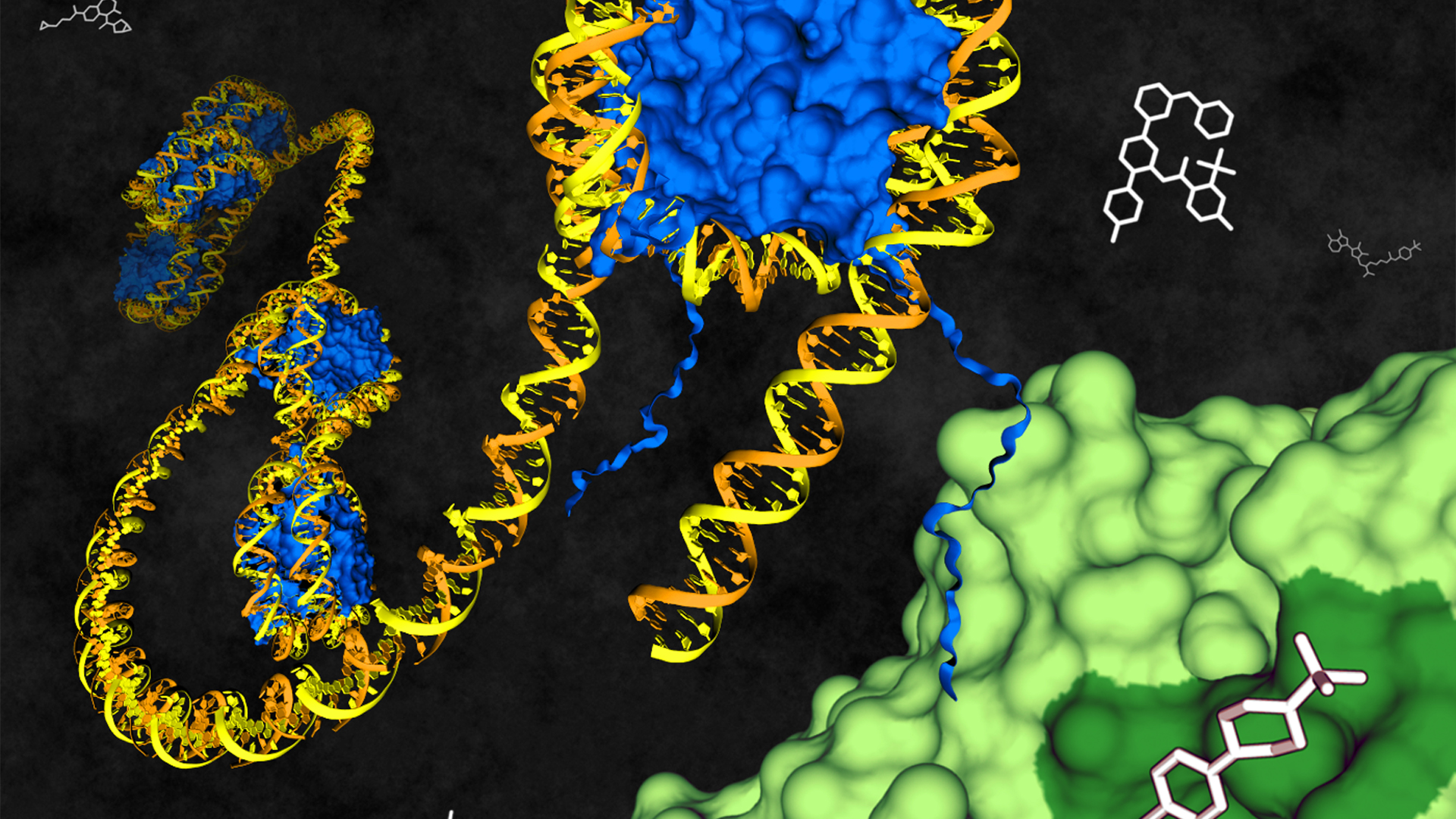
Targeting 20,000 proteins by 2035
When you run a web search for "Target 2035," most of the results are investment options. Aled Edwards and Cheryl Arrowsmith are hoping that by the time Generation X has begun to retire, a second meaning will be as common — at least in the chemical biology community.

Target 2035 is the name of their effort to inspire the community to develop a probe, or a specific small-molecule modulator, for each of humanity's 20,000 proteins.
The two Canadian structural biologists, among the leaders of the international Structural Genomics Consortium, are the driving force behind Target 2035, which launched in November.

Consortium
"It's an ambitious project," said pharmacologist Bryan Roth, who is not part of the consortium but has advised for it, a role he compares to reviewing for a journal. "It'd be great if we had real, useful chemical tools for all the targets in the genome."
Assessing the human genome project
Edwards began to consider the whole proteome about a decade after the Human Genome Project was completed in 2001. That project had identified 20,000 protein-coding genes, many of them for the first time.

of the Structural Genomics Consortium in Toronto
After some fellow researchers in the Structural Genomics Consortium in 2010 urged the cancer research community to explore as-yet-undrugged kinases instead of revisiting ones that already had been chemically validated, Edwards and collaborators conducted a literature review to see how research into other protein families, such as ion channels and nuclear receptors, had progressed. They found that most research had continued to circle historical targets.
"This makes no bloody sense!" he said. "If you were a logician and not a scientist, you'd say, 'Well, that's kind of dumb.'"
But science funding agencies and journal editors tend to reward a more conservative approach. Arrowsmith saw that while serving on a Canadian study section recently. "Everybody else on the panel said, 'Why (is the applicant) studying this protein? Nobody works on this protein; it doesn't do anything important that we know of.' I was disappointed, but it was kind of hard to argue."
During a recent seminar, Edwards said, "It's so much easier to work where people know about stuff. It's really hard to develop hypotheses about something that is unknown."
In their literature review, Edwards and his team also noticed a few proteins that had bucked the trend. Over the 10-year period they examined, those molecules had gone from obscurity to popularity. In those cases, Edwards said, almost without fail there was a potent and specific inhibitor available for the protein. Having a pharmacological approach to perturb a protein's activity made it much easier to study.
"We thought, OK, then why don't we start to proactively make research tools for proteins that no one currently gives a shit about?"
The SGC
The "we" was the Structural Genomics Consortium.

Founded by the Wellcome Trust, several Canadian research organizations and GlaxoSmithKline in 2004, the SGC is a nonprofit public–private partnership that aims to coordinate and accelerate protein structure discovery.
"Al was recruited to be the CEO of SGC, and the first person he asked to join him was me," Arrowsmith said. She became the chief scientific officer of the consortium's Toronto location.
Edwards and Arrowsmith, who met as structural biology trainees at Stanford University, already had collaborated on large-scale approaches to find protein structures as professors at the University of Toronto before getting involved in the nascent project.
Both said that their working styles complement each other: Edwards, an energetic speaker whose tangents sometimes eclipse his original point, appreciated Arrowsmith's focus and follow-through. She appreciated his big ideas.
"We kind of feed off each other in terms of what can be done and what should be done," Arrowsmith said.
The SGC now supports university-affiliated scientific teams in six countries and is funded by eight multinational pharmaceutical companies, Wellcome and Genome Canada.

"Initially it was purely a structure-determination organization," said Susanne Müller–Knapp, a molecular biologist who co-authored the initial kinase study and today coordinates activities at the Frankfurt arm of the consortium.
Structures and inhibitors can be mutually beneficial; knowing a protein's structure can help in the rational design of a small molecule to bind to it, while having a small molecule that binds to the protein can sometimes stabilize it, making crystallization and structural studies easier.
Over time, the group collected inhibitors of proteins they were studying and noticed when inhibitors were lacking. Müller–Knapp said, "At some point, we were wondering, 'We have these freezers full of proteins; what else can we do with them?'"
That was when the team started to work on developing small-molecule tools to bind to those proteins and block their activity, with a tight focus on epigenetic modulators, enzymes that modify DNA or histones or detect these modifications, to alter gene expression patterns.
Rigorously defining a probe
Chemogenomics
Even if a probe is not perfectly selective, it can be a useful tool for research. An approach called chemogenomics is one application.
Although genetic techniques to manipulate protein expression are well developed, the SGC team is not interested in generating 20,000 knockout mice or cell lines. They argue that removal of a protein through genome editing may have wider-reaching effects than blocking its function with a small molecule and is harder to translate into potential drug development.
Instead, the team focuses on small-molecule and biological tools, or probes, to block protein activity.
"Probe is a word that anyone can apply to their molecule," Edwards said during a webinar. But the SGC team applies a stringent definition: A probe is a molecular tool that acts selectively, by a known mechanism, to modulate protein activity.
Unlike a drug, a probe doesn't need to make its way through an organism to its target. Unlike an inhibitor, a probe doesn't need to block enzymatic activity; it might act, for example, by inducing protein degradation or binding to an allosteric site.
In some cases, a bad chemical tool could be worse than no tool at all. Arrowsmith cited a compound called DZNEP, often reported as an inhibitor of the widely studied histone methyltransferase EZH2. However, the compound actually was developed to disrupt the synthesis of S-adenosylmethionine, the cell's universal methyl group donor, wreaking havoc on every methyltransferase.

The original report on DZNEP was not strictly wrong, Arrowsmith said. "It does inhibit EZH2. It inhibits everything." However, interpreting experimental results after DZNEP treatment as specific to EZH2-mediated methylation would be a mistake. Unfortunately, Arrowsmith added, although this molecule's drawbacks have been known for some years, many reagent supply companies still offer it as a specific inhibitor of EZH2. "Even now, I still see papers where people use this DZNEP, which is a sloppy compound."
DZNEP is similar to other molecules known in the chemical biology community as pan-assay interference compounds, or PAINs. They act by a variety of mechanisms, but the uniting feature is that they are not specific to individual proteins. They tend to show up in the literature again and again, particularly after high-throughput screens, frustrating experienced medicinal chemists and leading less experienced ones astray in interpreting their data.
To be sure that each probe the SGC develops or distributes is selective for the target it was designed to block, the team collects a standardized set of assays. They look for data that demonstrate a probe binds to its target protein and blocks its activity in test tubes and in cells — and ideally can be cocrystallized, giving structural proof that it binds. They consider a molecule's selectivity between closely related proteins and its stability in solution to make sure it will not degrade into inactive compounds. Ideally, along with each probe, the SGC also hopes to distribute control compound that is chemically related but does not affect the same protein target. Two expert panels, one from within the SGC and one of external academics, review each probe to certify it before publishing.
Although the team has been successful in developing probes for epigenetics, the rigorous development process takes time. To achieve their genome-scale ambitions, the SGC team will need reinforcements. Edwards said, "We've spent considerable time and effort making 100 probes — and obviously, that won't scale."
The future of probe design
Computational techniques such as structure prediction and molecular docking could help researchers reach the 20,000 probe target.
Pharmaceutical partners
The largest developers of protein inhibitors are pharmaceutical and biotech companies, which routinely bombard protein targets with potential inhibitors and fine-tune the results one functional group at a time on the hunt for future drugs.
"Discovering new medicines is extremely difficult and also increasingly expensive," said Adrian Carter, the head of discovery research coordination at pharmaceutical company Boehringer Ingleheim, at a recent Target 2035 webinar. "Much of that high cost is driven by failure, unfortunately."
The SGC hopes that molecules that don't make it as drug candidates still might be useful as chemical probes — and that companies will make them freely available to researchers. This historically has been a difficult sell for companies whose business model depends on proprietary molecules. However, Arrowsmith said that thanks to a culture shift in the pharmaceutical industry, companies are now more willing to release molecules from "both past and failed drug-discovery programs where there are good modulatory molecules. Normally they'd just go sit on the shelf and wouldn't be used anymore. Now many companies are making these available to the community."
GlaxoSmithKline was one of the first companies to make samples and annotation about its probes openly available. In 2014, inspired by SGC scientists' work, the company made 367 previously published kinase inhibitors available to researchers.
In an article in the journal SLAS Discovery, a team of medicinal chemists originally from GlaxoSmithKline but now affiliated with the SGC described the process of publicizing those probes, which they called "extreme open science." First, the chemists collected the hundreds of inhibitors and screened each molecule for effects on more than 260 human kinases (see sidebar: Chemogenomics). Next, an equally daunting task, they worked out a process with the company's legal team to distribute molecules for research use under a simplified materials transfer agreement.
The kinase inhibitor set broke a trail for other companies to make their probes publicly available as well. Müller–Knapp led a team that recruited about 90 more selective molecules from numerous companies in a collection called the donated chemical probes panel, which they announced in a paper in eLife in 2018. Each compound had taken medicinal chemists years to develop, at a cost of up to 2 million euros apiece.

According to Müller–Knapp, companies are motivated to release the right to use their compounds in part to make the literature they rely on more reliable and reproducible. "If academics use compounds that are not characterized, they will always ascribe the phenotype to the so-called 'specific target' that the inhibitor was made for," she said. "And so the whole literature is polluted."
By enabling academic researchers to make more discoveries, these probes (which, after all, already had been rejected as drug candidates) could contribute to the pool of reliable knowledge about biology that industry researchers draw from regularly.
Besides, Arrowsmith said, making a small molecule available for research use is not the same as giving up the exclusive right to sell it. "If they let one of the molecules out there, they still may be covered by a patent, but they're making it available to the world to use."
But it's not without effort for companies. In industry, the discovery process is aimed at bringing molecules to market. If a probe does not have a reasonable path forward, researchers often stop experimentation. This leaves some molecules incompletely characterized by SGC standards. According to Müller–Knapp, companies that have committed to donating a set number of probes have occasionally dropped one and proffered another if the original candidate needed too much additional wet lab characterization.
Target 2035
The SGC has sent out thousands of samples of the epigenetic probe collection its scientists developed, the Published Kinase Inhibitor Set and the donated chemical probes set, according to Arrowsmith.Phil Cole, a pharmacologist at Harvard University who studies epigenetic modulators and has used some of the epigenetic probes, said, "SGC has had enormous impact in producing important protein structures and small-molecule probes and is one of the great success stories in big science applied in chemical biology."
SGC organizers say they're not sure whether the consortium will continue to function as a warehouse for the chemical probes they certify. To do so might institutionalize the SGC alongside nonprofits like Addgene, Jackson Labs and the American Type Culture Collection, or ATCC. On the other hand, Arrowsmith said, most SGC investigators are also active researchers and want to focus on using the probes they've worked so hard to develop to study biological questions.
Target 2035 kicked off with a series of webinars in November, and organizers have continued to hold monthly webinars on topics in drug development.
In the project's first phase, set to last until 2024, organizers aim to solicit donations of existing small molecules and develop processes for validating the molecules and sharing characterization data. In later phases they will try to use those data mining and biochemical profiling tools to coordinate and speed up probe development assays.
Like the improvements in sequencing during the Human Genome Project, Arrowsmith said, she expects the available tools for drug design to improve during the project, accelerating discovery.
How did they settle on the 15-year target date? "That was Aled," Arrowsmith said with a chuckle. "He pulled that out of a hat somewhere."
To achieve the goal of disrupting every protein in the proteome, Edwards explained, "You need to improve the technologies … You need to get better at chemistry, faster and more effective. But it doesn't break any laws of physics, right? So it should happen. It's just a matter of when. We chose 15 years as an aspirational goal and said, 'Let's go for it.'"
Most other scientists are skeptical about whether 2035 is a realistic end date. But pharmaceutical industry commentator Derek Lowe wrote, "The good news is that this isn't one of those efforts that has to make it to the end to be really valuable."
Target 2035 overlaps with a major National Institutes of Health initiative called Illuminating the Druggable Genome, which targets kinases, ion channels and receptors, and a more recent European effort called EUb Open that takes aim at 1,000 proteins. SGC adviser Roth said that because of differences in funding streams, "They're all separate projects, sadly. It'd be great if we could all be part of one gigantic project."
According to Edwards, bringing major international projects, industry groups and individual academic labs into alignment is precisely the goal of Target 2035. "I'm talking about a science project," he said. "But my No. 1 and 2 projects are culture."
Machine learning and the future of probe design
In November, the protein folding program AlphaFold made headlines by correctly predicting the structures of some two dozen proteins that had been solved by researchers but were unpublished.
"Bingo! Miracle result," Edwards said. "Only 50 years in the making."
The folding program evolved from a structural biology competition called CASP, or Critical Assessment of Structure Prediction, which organizers always had envisioned as a training ground for new structure prediction tools. Using sequence–structure relationships made public in the Protein Data Bank as a training set, the competition has challenged participants to extrapolate from a known sequence to a new structure.
A success like AlphaFold's "was exactly the plan of structural biologists," Edwards said. "They've spent literally billions and billions of dollars creating the foundation so that this could happen."
Just as the Protein Data Bank collected and organized knowledge about protein structure, becoming an integral resource for machine learning, Edwards said that the Structural Genomics Consortium, which he co-founded, means to organize knowledge about chemical probes and their activities that may be used for data mining in the future.
Researchers already are beginning to use computational tools to develop new probes. For example, in a Nature paper in 2020, labs at the University of North Carolina and University of California San Francisco used computational tools to dock 150 million hypothetical compounds to a melatonin receptor, synthesizing and screening only the most promising.
The approach dramatically expands the speed of screening, said Bryan Roth, who led the UNC team. "It's now possible, once you have a structure of a protein, to dock billions of small molecules that don't actually exist in the physical universe … then to get them synthesized, and to develop probes that way."

Chemogenomics: Putting imperfect probes to use
Finding a selective modulator of a protein can be difficult, according to Cheryl Arrowsmith, a structural biologist and co-founder of Target 2035. "Often, one has to make a compromise with saying, 'okay, this compound is good enough; let's publish it and see what we can do with it, warts and all, if you will,'" she said. "The issue is, everybody should know what the warts are."
Some approaches take the warts into account and work around them. For example, adenosine triphosphate mimics tend to block numerous kinases. An approach developed by chemists at the Structural Genomics Consortium and numerous pharmaceutical companies involves applying many kinase inhibitors, each of which targets a relatively small number of kinases, and using the overlap in results to determine exactly which kinase target is responsible for an assay result. The approach is called chemogenomics.

Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Building the blueprint to block HIV
Wesley Sundquist will present his work on the HIV capsid and revolutionary drug, Lenacapavir, at the ASBMB Annual Meeting, March 7–10, in Maryland.
Gut microbes hijack cancer pathway in high-fat diets
Researchers at the Feinstein Institutes for Medical Research found that a high-fat diet increases ammonia-producing bacteria in the gut microbiome of mice, which in turn disrupts TGF-β signaling and promotes colorectal cancer.

Mapping fentanyl’s cellular footprint
Using a new imaging method, researchers at State University of New York at Buffalo traced fentanyl’s effects inside brain immune cells, revealing how the drug alters lipid droplets, pointing to new paths for addiction diagnostics.

Designing life’s building blocks with AI
Tanja Kortemme, a professor at the University of California, San Francisco, will discuss her research using computational biology to engineer proteins at the 2026 ASBMB Annual Meeting.
Cholesterol as a novel biomarker for Fragile X syndrome
Researchers in Quebec identified lower levels of a brain cholesterol metabolite, 24-hydroxycholesterol, in patients with fragile X syndrome, a finding that could provide a simple blood-based biomarker for understanding and managing the condition.
How lipid metabolism shapes sperm development
Researchers at Hokkaido University identify the enzyme behind a key lipid in sperm development. The findings reveal how seminolipids shape sperm formation and may inform future diagnostics and treatments for male infertility.

