Lively lysosomes
Lysosomes are having a Cinderella moment. Gone is the perception that lysosomes simply sit in a corner of a cell, mutely cleaning up whatever is sent their way. Lysosomes now are seen as critical signaling checkpoints that move around the cell and take part in important decisions about cellular biosynthesis and degradation. They once were thought to be involved in only rare genetic disorders. But now researchers are beginning to appreciate that diseases as common as Alzheimer’s and certain cancers have roots in lysosomes too.
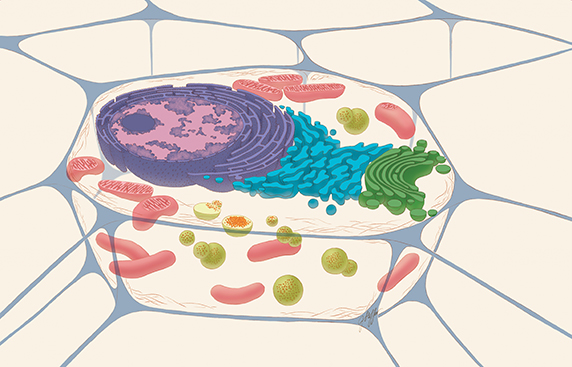 A typical drawing of an animal cell, sliced open to reveal cross-sections of organelles. The lysosomes are the green spheres. The red dots within the green spheres signify cellular components that need to be broken down.THE NATIONAL INSTITUTES OF HEALTH
A typical drawing of an animal cell, sliced open to reveal cross-sections of organelles. The lysosomes are the green spheres. The red dots within the green spheres signify cellular components that need to be broken down.THE NATIONAL INSTITUTES OF HEALTH
Along with the reassessment of the capabilities of lysosomes comes the awareness that scientists have more questions than answers about exactly what the organelles are capable of doing. Lysosomes, says Roberto Botelho at Ryerson University in Canada, “are much more fun than people thought.”
Trash can
In 1955, Christian de Duve at the Catholic University of Louvain in Belgium and colleagues described a membrane-bound organelle that housed at least five enzymes. These enzymes degraded a variety of substrates in a pH around 5. In proposing that the organelle was involved in cellular digestion, de Duve and colleagues called the organelle a “lysosome,” the Greek word for “digestive body.”
In subsequent years, researchers found that there are at least two ways for molecules to wind up in one of the hundreds of lysosomes in a cell. One way involves endocytosis, in which molecules outside of the cell are brought inside the cell in packages. Some of the packages are fated to become late endosomes, which are slightly acidic organelles that mature into lysosomes.
Another way is autophagy. This is a major housekeeping mechanism within the cell, clearing away components that are about to expire. The cleared components arrive at lysosomes in vesicles known as autophagosomes.
Once molecules are in a lysosome, nucleases, proteases, lipases and other hydrolytic molecules attack them. Exporters on the membrane carry out the bits and pieces of the degraded molecules. The pieces go into the cytoplasm either to provide energy or to be reused by the biosynthetic pathways.
Lysosome, not lysozyme
 De Duve
De Duve
In an article published in Nature Cell Biology on the 50th anniversary of the lysosome’s discovery, Christian de Duve was peeved with scientists who confused “lysosome” with “lysozyme,” a bacterial enzyme discovered by Alexander Fleming of penicillin fame. “I trusted biochemists to be able to distinguish between the Greek roots soma and zyme,” he wrote in the 2005 perspective. “This trust was sadly misplaced. Even today, I am still sometimes given credit that is due to Fleming.”
In 1963, Henri-Gery Hers, who had joined de Duve’s group, discovered that people missing a lysosomal glucosidase succumbed to a severe glycogen storage disorder. That finding introduced the idea that a host of diseases could be linked to the inability of the lysosome to produce specific enzymes and thus degrade particular molecules. Lysosomal research at that point became focused largely on the clinical aspects of disorders associated with the lysosomes and finding therapies for the disorders.
Disorders related to the inability of the lysosome to break down and remove various types of molecules became known as lysosomal storage disorders. These included Gaucher, Fabry and Niemann–Pick diseases. There are now more than 50 known lysosomal storage disorders, most of them rare genetic diseases.
One of the notable breakthroughs in this area was enzyme-replacement therapy, during which certain missing or defective lysosomal enzymes are replaced with functional enzymes, as Roscoe Brady’s team at the National Institutes of Health did with Gaucher disease.
By the 1980s, it seemed scientists knew what they needed to know about the basic workings of the lysosome. “Who wanted to work on lysosomes? It was more interesting to work on the nucleus, where all the genetic information is contained, or mitochondria that make energy for the cell to function or the (endoplasmic reticulum), where proteins are synthesized,” says Juan Bonifacino at the NIH. “Lysosomes were just involved in degradation. They were a trash can.”
Then came an unexpected finding.
‘They didn’t like the idea’
David M. Sabatini at the Whitehead Institute is the first to admit that he should have listened to his father’s advice. When Sabatini was in graduate school at Johns Hopkins University in the early to mid-1990s, he identified a kinase that is targeted by an immunosuppressant drug called rapamycin. That serine-threonine kinase is the mammalian target of rapamycin, or mTOR. Scientists soon found mTOR to be a critical player in cellular growth and implicated in a number of cancers.
mTOR comes in two complexes. One, mTORC1, is exquisitely tuned to amino acid levels in the cell. Researchers showed that the presence of amino acids triggered the activation of mTORC1. But how and where the kinase checked in on the amino acid levels was a mystery.
Sabatini is a second-generation scientist. His father, David D. Sabatini, is a cell biologist at New York University. “When I first identified mTOR as a graduate student, I remember I was talking to (my dad) about it. He said, ‘David, one of the things you have to do is you have to localize this within the cell,’” recalls the younger Sabatini. “I was a typical obnoxious child, and I was like, ‘You know, I don’t think that’s interesting. That’s old school.’ The funny thing is that it turned out that the localization was the key thing.”
In 2008, nearly a decade after he began working with mTOR, the younger Sabatini, who also is with the Howard Hughes Medical Institute, led a team that made a surprising discovery. When the team deprived cells of amino acids, mTORC1 was diffuse throughout the cytoplasm. When the team added amino acids, the kinase quickly congregated on the surface of lysosomes.
“I remember the first few times I presented this finding, people would stand up and say that the lysosome was a trash can. Some people would be more charitable and say the lysosome was a recycling bin,” recalls Sabatini. “They didn’t like the idea. The finding was met with a bit of resistance because it was one of the first that implicated there was something different about lysosomes.”
The resistance turned to curiosity when Sabatini’s group published the finding in the journal Science later in 2008. A critical kinase that oversaw cell growth was making the lowly lysosome its headquarters when activated.
Sabatini “is single-handedly responsible for putting the mTORC1 signaling complex on the lysosomal membrane,” says Michael Overholtzer at Memorial Sloan Kettering Cancer Center. “That really brings the lysosome to the forefront.”
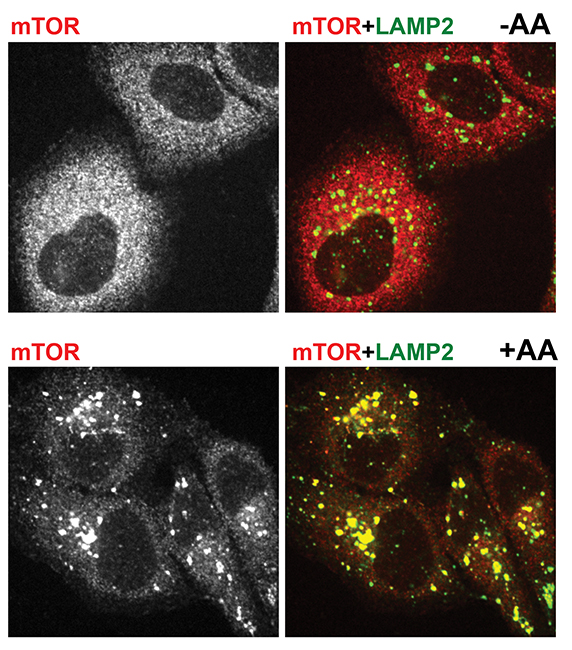 Immunofluorescent pictures show mTOR in red and a lysosomal marker in green. The top two pictures are from amino acid-starved cells. mTOR is dispersed and does not localize with lysosomes. The bottom two pictures are from cells that were starved and then given amino acids. There mTOR clusters on lysosomes.ROBERTO ZONCU AT THE UNIVERSITY OF CALIFORNIA, BERKELEY
Immunofluorescent pictures show mTOR in red and a lysosomal marker in green. The top two pictures are from amino acid-starved cells. mTOR is dispersed and does not localize with lysosomes. The bottom two pictures are from cells that were starved and then given amino acids. There mTOR clusters on lysosomes.ROBERTO ZONCU AT THE UNIVERSITY OF CALIFORNIA, BERKELEY
Clearly not a dead end
The next indication that the lysosome wasn’t a mere refuse receptacle came in 2009 from Andrea Ballabio’s laboratory at Telethon Institute of Genetics and Medicine in Italy. His group published a paper in Science that showed that lysosomal genes are regulated by a single protein called transcription factor EB, or TFEB.
Researchers knew that a single cell contains hundreds of lysosomes, enough to make up about 5 percent of the cell’s volume. But they thought the number stayed the same over the course of a cell’s lifetime.
Ballabio and colleagues suspected otherwise. “We postulated that any cell needed to have a mechanism to modulate lysosomal function,” he says. “This was actually a relatively new way of thinking, because the traditional view of the lysosome was of a static organelle not subject to regulation and adaptation. But we postulated that there was a network of genes encoding for lysosomal proteins that would be jointly regulated” by a common entity.
Ballabio and colleagues analyzed the expression of genes encoding lysosomal proteins under multiple conditions and situations. And they did so without picking up a pipette.
“We didn’t even do the experiments ourselves because the experiments were out there in the databases,” says Ballabio. “We looked at microarray databases, where there are experiments done under many different conditions, and looked at all known genes encoding lysosomal proteins.”
Ballabio’s team discovered that the expression levels of the lysosomal genes went up and down in a coordinated fashion. When they looked at the promoter regions of the lysosomal genes, they found that there is a common sequence, the CLEAR site, in many lysosomal gene promoters. This site was a known target site for the transcription factor TFEB. Shortly thereafter, Ballabio’s group found that TFEB regulates autophagy, which implicated TFEB in controlling both cargo delivery to the lysosome and degradation.
Deciding factor
Then the worlds of TFEB and mTORC1 collided. In 2012, the groups of Sabatini and Ballabio demonstrated that TFEB and mTORC1 show up on the same spot of the lysosomal membrane. When nutrients are abundant in the cell, mTORC1 phosphorylates TFEB and keeps it inactive on the lysosome. When nutrients, such as amino acids, drop in abundance, mTORC1 becomes inactive and no longer phosphorylates TFEB. The unphosphorylated and active transcription factor takes off for the nucleus to turn on lysosomal genes and turn up the cell’s degradative capabilities to either reshuffle allocation of materials or provide energy.
With the finding that TFEB and mTORC1 partner, says Overholtzer, it is obvious that “the lysosome is not simply a dead end.” It directly communicates with the nucleus, the cell’s main control center, and partakes in decisions about growth and degradation.
‘Much more complicated’
The lysosome’s signaling roles appear to be even more sophisticated than first thought. Haoxing Xu at the University of Michigan, Ann Arbor, leads a research team studying a class of calcium channels found spanning the lysosomal membrane. Mutations in the channels, called mucolipin TRP proteins, cause a rare neurodegenerative disease in children.
Xu says there is some evidence that when the amino acid levels are low and mTORC1 is not active, the calcium channels kick into action, releasing calcium, an important signaling ion, from the lysosome into the cytoplasm.
This lysosomal calcium signaling regulates TFEB. Ballabio’s group discovered that during starvation, lysosomal calcium release activates a phosphatase that dephosphorylates TFEB. The dephosphorylated TFEB moves into the nucleus to kick off more lysosome biogenesis and autophagy.
Taken together, mTORC1, TFEB and the calcium channels “constitute a signaling network to regulate when the degradation should occur and when degradation should be terminated,” says Xu. “It’s much more complicated than previously thought.”
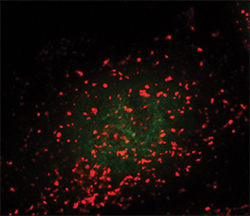 Lysosomes are seen as the red dots. The green dots show active TFEB in the nucleus.ANDREA BALLABIO AT THE TELETHON INSTITUTE OF GENETICS AND MEDICINE
Lysosomes are seen as the red dots. The green dots show active TFEB in the nucleus.ANDREA BALLABIO AT THE TELETHON INSTITUTE OF GENETICS AND MEDICINE
On the move
It’s becoming abundantly clear that lysosomes are not one-trick ponies. For example, they are capable of repairing the plasma membrane.
In 1997, Norma Andrews’ group, now at the University of Maryland, showed that lysosomes can function as calcium-regulated secretory vesicles. They don’t just take things in; they are capable of releasing molecules. The finding was met with a lot of resistance at the time, since “conventional lysosomes were not expected to do that,” says Andrews.
In 2001, the group moved their findings further along by demonstrating that the lysosome responds to calcium entering through tears in the plasma membrane by fusing with the boundary to heal it. The calcium-controlled process is known as lysosomal exocytosis.
But for exocytosis to happen, lysosomes need to move. As researchers now appreciate, lysosomes don’t just sit in a spot. Depending on conditions in and surrounding a cell, lysosomes move back and forth between the center and the periphery of the cell. They do so by coupling to microtubule motors, kinesin and dynein through an elaborate set of adaptor molecules, says Bonifacino, adding that the attachment to motors “allows the lysosomes to patrol the whole cytoplasm, looking for places where they can exert their activity.”
But the movements and duties of the lysosome can be hijacked, explains Andrews. That’s exactly what the protozoan Trypanosoma cruzi, which causes Chagas disease, is capable of doing. The pathogen recruits lysosomes to the plasma membrane, makes them fuse with the plasma membrane and tricks them into reforming with the parasite in them. When the lysosomes travel back to the cell interior, the parasite burrows out of the organelles and takes over the cell.
Indeed, defects in lysosomal movement are implicated in disease. If lysosomes are forced to be immobile in a normal cell, “a lot of things go wrong in the cell,” notes Bonifacino. He gives the example of autophagy. If lysosomes are forced to hold still, autophagosomes build up without having lysosomes nearby to fuse with. This spells trouble for the cell.
Cell migration and adhesion also rely on moving lysosomes. “Late endosomes or lysosomes move to sites of cell adhesion or migration, and they bring adhesion molecules and signaling molecules like mTOR or MAP kinases that remodel those adhesive structures,” says Bonifacino. “That allows the cell to move. If you inhibit lysosome motility specifically then the cells become less mobile.”
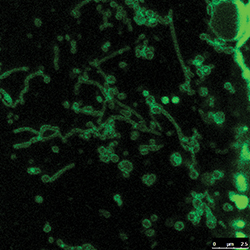 Not much is known about the long, snakelike tubulated lysosomes.HAOXING XU AT THE UNIVERSITY OF MICHIGAN
Not much is known about the long, snakelike tubulated lysosomes.HAOXING XU AT THE UNIVERSITY OF MICHIGAN
New questions
The renewed interest in lysosomes brings with it new questions. For example, researchers know that lysosomes in certain cell types, such as the immune cell’s macrophages, can be long and snakelike. In other cells, lysosomes tend to be round sacs, ranging from 100 to 1,000 nanometers in diameter.
One tantalizing question: Is there a difference between tubular and round lysosomes? In the case of the macrophages, the cells that engulf and destroy all kinds of unwanted matter, the snake shape is thought to help the lysosomes better pass on peptides from unwanted matter to the plasma membrane so that the immune system knows which entities to search for. But, as Ryerson University’s Botelho stresses, “Very, very little is known about tubular lysosomes.”
Very little also is known about the physical organization of the lysosome. Researchers estimate there are more than 50 types of enzymes inside the lysosome. Do these enzymes flit about like attendees at a cocktail party? Or are they assigned to specific places like workers in a factory? No one knows.
Even for something as critical as mTORC1, the details are hazy. So far researchers know that there are small GTPases that physically anchor mTORC1 to the lysosome. The GTPases have their own set of regulators that in turn are controlled by a proton pump, the vacuolar ATPase, which maintains the acidic pH of the lysosome interior. But how exactly is mTORC1 detecting amino acid levels, the job that it’s known for doing?
Sabatini says his group has identified a protein that appears to transmit information about amino acid levels from the lysosome interior to mTORC1. But, he adds, they are working on proving that what they have found is a bona fide amino acid sensor.
Extending the reach of lysosomes
It’s not just nit-picky molecular questions that are coming up about lysosomes. There are some fundamental questions too. For example, are all lysosomes the same?
“It’s very likely that the composition of the lysosome changes from organ to organ depending on the specific metabolic needs,” says Roberto Zoncu at the University of California, Berkeley, who was the postdoctoral fellow in Sabatini’s group when it spearheaded the mTORC1 localization. “For example, in the liver, you have a lot of glycogen production and storage. It’s possible that lysosomes might be specialized in handling sugars.” But, he adds, this aspect of lysosomes is not well-explored.
Researchers are wondering if, even within a single cell, there are differences within the hundreds of lysosomes. Different groups of lysosomes can be tasked with different jobs in the cell. For example, Andrews says, “my prediction would be there is a population specialized in associating with the plasma membrane and is involved in plasma membrane repair.”
Another big question is about the influence of the lysosome on an entire organism. “Just how far does this system go? If the lysosome is a signaling hub, what is the full range of actions that it can have on the body?” says Zoncu. If the lysosome plays critical roles in signaling cell growth and degradation, how do these roles play out on whole-body parameters, such as growth and metabolism?
Researchers also are now very interested in studying the lysosome’s roles in diseases such as neurodegeneration and cancer. “Some cancers upregulate their lysosomal complement massively,” says Zoncu. “There are several reports now showing that some cancer types, especially Ras-based cancers, are literally addicted to lysosomal functions. Whether this is a stress response pathway or if it’s a way for them to scavenge nutrients, I think this is a great direction of investigation.”
For neurodegenerative diseases, Bonifacino uses Alzheimer’s to illustrate how researchers are starting to think lysosomes may be involved. A signature of Alzheimer’s disease is an accumulation of plaques in the brain. The plaques are aggregates of a peptide called beta-amyloid, which is secreted from neurons and glial cells into the areas around the cells.
Although this extracellular beta-amyloid was long thought to be toxic to neurons, says Bonifacino, recent work suggests that beta-amyloid inside the cells may be to blame for causing neuronal damage. Here changes in lysosome function could lead to damaging accumulation of beta-amyloid inside cells.
The element of surprise is the continuous thread in lysosome research. Even the discovery of lysosomes happened as a tangent. De Duve’s group actually was chasing the action of insulin on the liver when it stumbled across the acidic digestive body in cell-fraction studies. Surprise after surprise came with associations with signaling molecules and other unexpected features of lysosomes.
So, these days, researchers no longer relegate lysosomes to the corner. They put lysosomes on center stage and continue to be beguiled.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Women’s health cannot leave rare diseases behind
A physician living with lymphangioleiomyomatosis and a basic scientist explain why patient-driven, trial-ready research is essential to turning momentum into meaningful progress.

Life in four dimensions: When biology outpaces the brain
Nobel laureate Eric Betzig will discuss his research on information transfer in biology from proteins to organisms at the 2026 ASBMB Annual Meeting.

Fasting, fat and the molecular switches that keep us alive
Nutritional biochemist and JLR AE Sander Kersten has spent decades uncovering how the body adapts to fasting. His discoveries on lipid metabolism and gene regulation reveal how our ancient survival mechanisms may hold keys to modern metabolic health.

Redefining excellence to drive equity and innovation
Donita Brady will receive the ASBMB Ruth Kirschstein Award for Maximizing Access in Science at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Mining microbes for rare earth solutions
Joseph Cotruvo, Jr., will receive the ASBMB Mildred Cohn Young Investigator Award at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Fueling healthier aging, connecting metabolism stress and time
Biochemist Melanie McReynolds investigates how metabolism and stress shape the aging process. Her research on NAD+, a molecule central to cellular energy, reveals how maintaining its balance could promote healthier, longer lives.

