A small army of researchers races
to build a coronavirus interactome
As institutions shuttered across the U.S. ahead of shelter-in-place orders, Nevan Krogan’s lab at the University of California, San Francisco, raced to gather the last bits of data they needed to assemble a protein interaction map to identify drugs that might work against SARS-CoV-2, the virus that causes COVID-19.
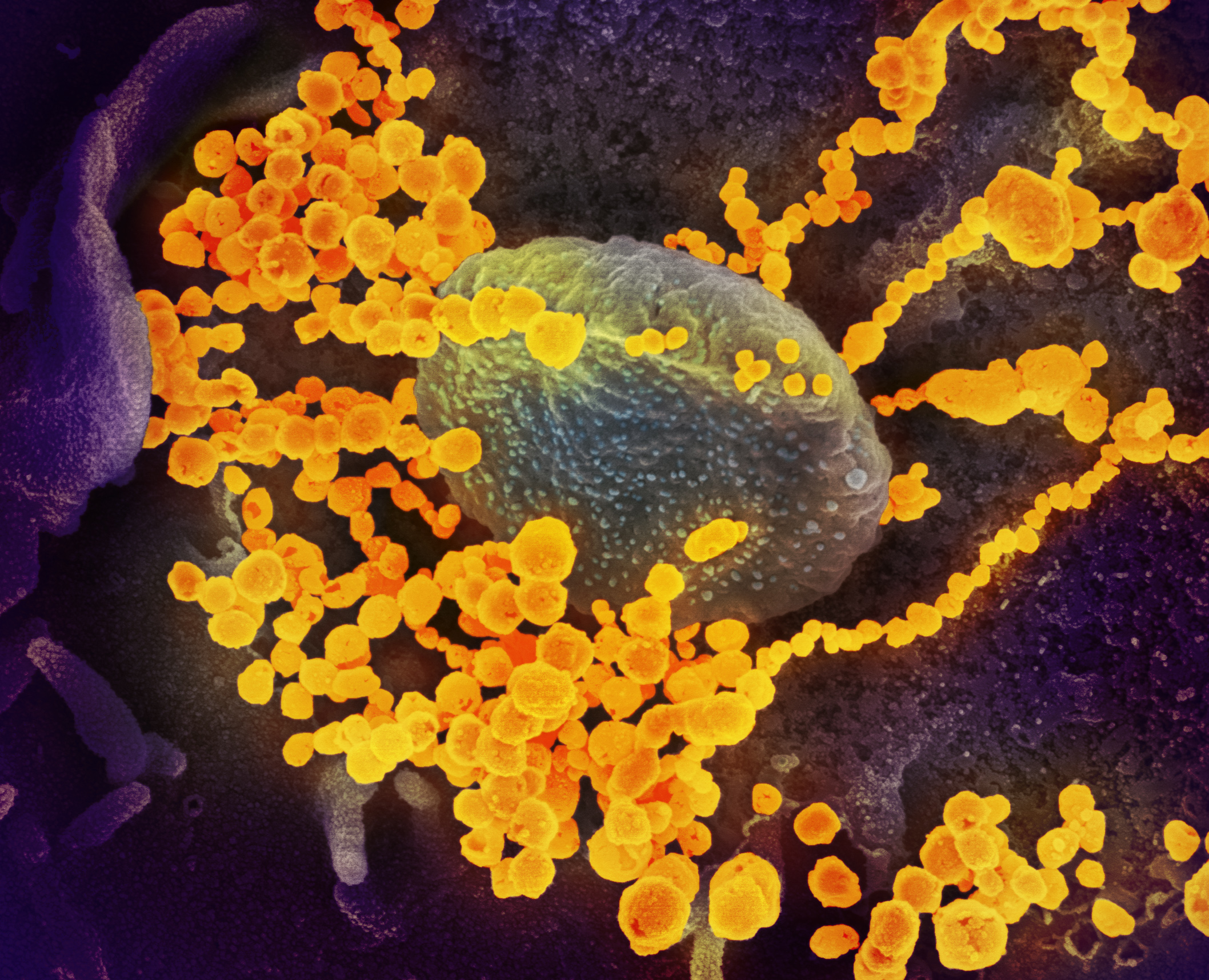
“We actually collected the last sample for the map about six hours before they shut everything down,” said Krogan, a professor in the department of cellular and molecular pharmacology at UCSF and the director of the university’s Quantitative Biosciences Institute. “It was a really a race against the clock, and it was a testament to the scientists in the lab essentially working for days on end without sleeping.”
In a study uploaded to the preprint site bioRxiv last week, Krogan and 93 co-authors list 69 drugs that might be effective in targeting COVID-19, the coronavirus that is testing the limits of healthcare systems and safety nets around the world. To identify these drugs, they built an interactome that showed how 332 human proteins interacted with proteins from 26 of the virus’ 29 genes.
“It’s a kind of a blueprint of how the virus comes in and hijacks and rewires the host during the course of infection,” Krogan said. “What happens is you put a hook on each one of (the viral genes) and you express the genes in human cells, and then what you do is pull out, fish out, the corresponding protein with that hook, and then you see by mass spectrometry which other human proteins are physically interacting or physically talking to the viral proteins.”
Krogan conceived and coordinated the collaborative effort, which was carried out by scientists from 22 labs at the Quantitative Biosciences Institute as well as from the University of Michigan, the University of California San Diego, the Icahn School of Medicine at Mount Sinai, the Howard Hughes Medical Institute, the Fred Hutchinson Cancer Research Center, the European Molecular Biology Laboratory and the Institut Pasteur. When it came to determining which drug targets could interact with the viral proteins, Krogan looked to UCSF chemists Brian Shoichet and Kevan Shokat.
“For the last 10 years, we've been working on getting drugs for undruggable targets,” Shokat said. “I looked in the map for weird targets that nobody thinks of as a traditional drug targets.”
That meant the target list included not only the immunosuppressant rapamycin and the drugs chloroquine and remdesivir, which are being evaluated against SARS-CoV-2 in clinical trials around the world, but also cancer drugs such as dabrafenib and natural products such as WDB002, a cyclic polyketide synthase, that are being evaluated for clinical use.
All of those drugs are being evaluated against the coronavirus in the biosafety-level 3 labs at the Institut Pasteur in Paris. There, Marco Vignuzzi and his lab members, including postdoctoral fellows Bjoern Meyer, Veronica Rezelj and Cassandra Koh, must work one or two at a time due to the city’s continuing lockdown.
“We are currently testing 20 compounds,” Meyer said. “Overall, I think the whole panel will include something along the lines of 65 or 69 compounds.” Their efforts are being duplicated, for the sake of corroboration, by Kris M. White and Lisa Miorin at the Icahn School of Medicine at Mount Sinai in New York.
The collaboration includes researchers at UCSF who hadn’t previously brought their skills to bear on viral problems, such as structural biologist Natalia Jura.
“We were just so motivated to contribute that everybody really put on hold what they were doing … and just started reading papers about coronaviruses and accumulating expertise in this area,” Jura said. “As a structural biologist, I was most interested in understanding what is known about how viral proteins interact with the host proteins.”
Most of the paper’s authors are now analyzing data remotely, with little access to their labs, according to Krogan.
“We're having regular Zoom calls with scientists — I think last time there were 100 different scientists on the Zoom call discussing this map,” he said. Krogan and colleagues at the UCSF-affiliated nonprofit Gladstone Institute previously assembled drug interaction maps for HIV and for the Ebola, Zika and dengue viruses with much longer timeframes.
“Normally, that takes a few years,” he said. “Well, we expedited this for a couple of weeks. And that's a testament in my opinion to the collaborative effort that was underway.”
Shokat is impressed by the speed with which the group was able to pull together its data.
“Usually we have six months, nine months to do it. But here you had two weeks,” he said. “It was really outstanding. You just wake up at 7, work till 11 and then check your phone until it's bedtime. It’s crazy, everybody's spirit has been amazing.”
The scientists in San Francisco must work at a distance from each other and from their labs, but their colleagues in New York and in Paris continue to evaluate compounds against the coronavirus in their level 3 labs, the second-highest level of biosafety containment. As Krogan and colleagues whittle down their list of drugs, he hopes that even more scientists will take advantage of their interaction map to find new leads about both SARS-Cov-2 and drugs that might work against it.
“It's a very rich data set, and they can make predictions about the biology of virus that we didn't find, and they can also make predictions about other drugs and compounds,” he said. “What I'm hoping for here, the silver lining in all of this, is that we're setting a new paradigm of how to do science. And hopefully, this infrastructure stays in place so that we're in a better position to tackle the next pandemic.”
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

Chemistry meets biology to thwart parasites
Margaret Phillips will receive the Alice and C. C. Wang Award in Molecular Parasitology at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

ASBMB announces 2026 JBC/Tabor awardees
The seven awardees are first authors of outstanding papers published in 2025 in the Journal of Biological Chemistry.
Missing lipid shrinks heart and lowers exercise capacity
Researchers uncovered the essential role of PLAAT1 in maintaining heart cardiolipin, mitochondrial function and energy metabolism, linking this enzyme to exercise capacity and potential cardiovascular disease pathways.

Decoding how bacteria flip host’s molecular switches
Kim Orth will receive the Earl and Thressa Stadtman Distinguished Scientists Award at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Defining JNKs: Targets for drug discovery
Roger Davis will receive the Bert and Natalie Vallee Award in Biomedical Science at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

