‘My involvement with ASBMB has made me want to do more’
Ann Stock, a professor at Rutgers University’s Center for Advanced Biotechnology and Medicine, became president of the American Society for Biochemistry and Molecular Biology on July 1. During the ASBMB annual meeting in April, Stock sat down for a conversation with ASBMB Today.

You can’t talk about Stock’s career without discussing her lifelong research interest in two-component systems, a large family of evolutionarily related signaling systems common in bacteria. As a graduate student, before the amount of sequence identity needed to establish homology had been well established, Stock helped to identify this protein family, finding weak sequence similarities between proteins encoded by an operon she had sequenced in Salmonella and other known bacterial sequences. She has continued to study two-component systems from different angles ever since.
Stock is a fellow of the American Academy of Microbiology and the American Association for the Advancement of Science; she’s a former Howard Hughes Medical Institute investigator and has served on the ASBMB Council and committees continuously since 2008. For more on her plans as president, see the President’s Message on page 2 of this issue.
This interview has been condensed and edited for clarity.
When did you decide that you wanted to be a scientist?
My initial interest in science came from my father. He actually never made it through high school; his father died during the flu pandemic of 1918, and he had to drop out to support his family, bagging groceries. That was the end of his formal education, but he loved natural history, geology, plants, visiting museums, watching National Geographic.
I started down a science track in college thinking that I would go into medicine. The pre-med advisors at University of California, Berkeley, said to volunteer at the hospital. I walked into the hospital and started down a hallway lined with patients on gurneys to get to the candy striper office. I never made it to the office.
People that go down the medical path have so much compassion, and I admire them enormously, but I realized right then and there that medicine was not for me. I went back to the university and took a job at the student learning center.
I managed to get into a lab in my senior year. Back then, biochemistry had three years of prerequisites. You’d knock on doors asking for research experience; everyone wanted to know, Have you taken the lab courses? I hadn’t. Finally, in my senior-year biochemistry course, I got along really well with my teaching assistant, Mark Snyder. He dragged me upstairs and knocked on the door of his advisor, Dan Koshland — who, to me, was like a god in the field. (Author’s note: Koshland, a former ASBMB president, was famous for his work in enzymology and in bacterial chemotaxis, for which he later received the Lasker award.)
I spent the latter part of my senior year doing research in Dan’s lab and stayed on for a year as a technician. During that time, I was teaching freshman chemistry extension courses in the evenings. I thought I wanted to go to graduate school in order to teach, but then I got bitten by the research bug and decided I wanted a position that balanced both.
You stayed on in the Koshland lab as a graduate student?
Yes. I got stuck with a project that they would stick only an undergraduate on, because it was deemed more or less undoable. When I transitioned to graduate school, I had a really hard time getting through my qualifying exam research proposal, because my committee thought the project would never work.
We were studying bacterial chemotaxis. My job was to find out the role of one of the proteins that seemed to be a master response regulator that controlled the direction of flagellar rotation.
Before molecular biology techniques were widespread, my approach was to purify the protein from wild-type cells using two-dimensional gels for isolation. I would run an isoelectric focusing gel in a tube, and then I’d push this little worm of a gel out of the tube, layer it on top of a regular SDS gel and seal it with agarose, and then run the second dimension. I managed to cut enough protein out of gels and used it to make antibodies in rabbits. It took gel after gel after gel. You can understand why my committee thought, “What if you don’t get antibodies after this heroic effort?”
Once we had antibodies, we had a handle on purifying the protein, which allowed us to begin to study it. I switched over to a molecular biology approach as that became more accessible. I sequenced an operon that encoded about half of the chemotaxis proteins. When I deposited it in GenBank, it doubled the amount of sequence data available for Salmonella typhimurium, as it was known then — now it’s Salmonella enterica.
We noticed weak sequence similarity between portions of two of the proteins and three other previously sequenced proteins that had nothing to do with chemotaxis but were involved in responses to osmolarity, dye uptake and sporulation. They had no apparent functional relationship to each other. Back in the day, people were used to thinking about homologous proteins as having sequence identity of 80% to 90%. No one thought that 20% to 30% sequence identity was significant. And I struggled to get the story out.
This was back in a time when a member of the National Academy of Sciences, like Dan Koshland, could just submit a paper to the Proceedings of the National Academy of Sciences. We submitted the paper, and then I presented my thesis and made what I thought was a wonderful case that these proteins were all related in terms of carrying information to elicit a response. My committee looked at me like I was crazy. Dan got such cold feet he pulled the paper.
So what happened?
He sat on it for a while. We continued to convince him. And then the paper was resubmitted.
Did you have to do many more experiments?
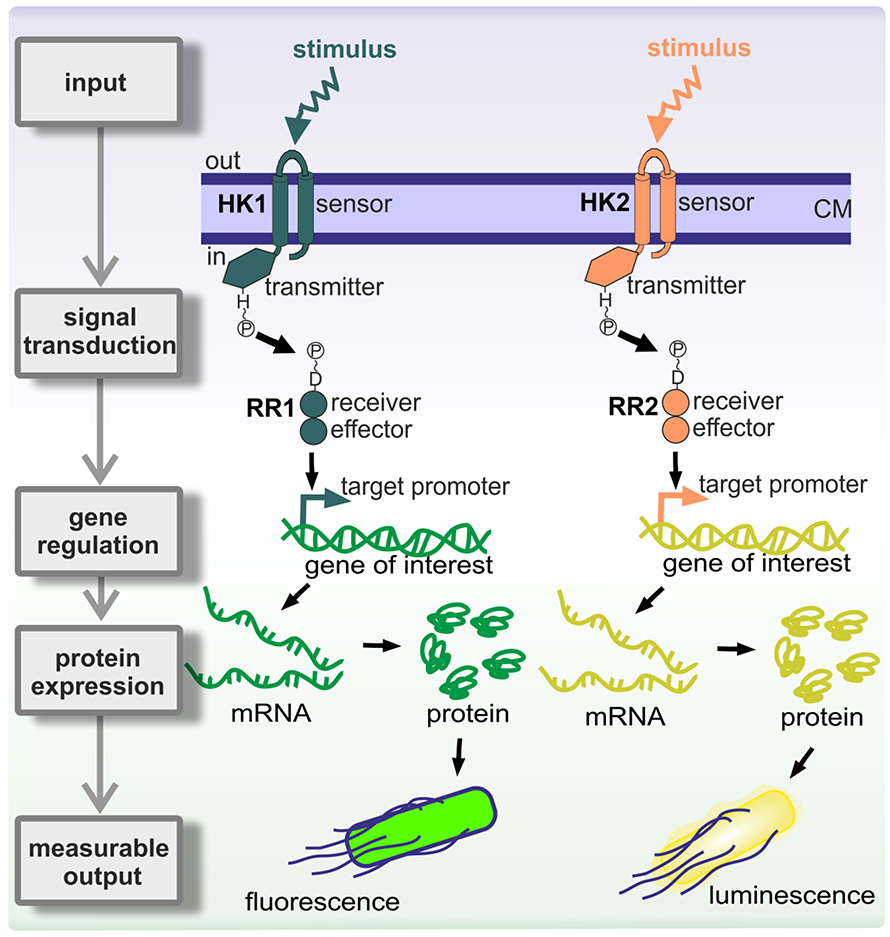
No, actually. It was really just talking it through. I guess a few things were starting to come out; Dan was hearing more and more within the field that there might be some links between these homologous proteins. Anyway, this established the family of response regulators. Soon after, these proteins were found always to exist in systems along with one other protein from a family that had a similar 20% to 30% sequence identity. Because there were two proteins that were found together, they were called two-component signaling systems — a terrible name.
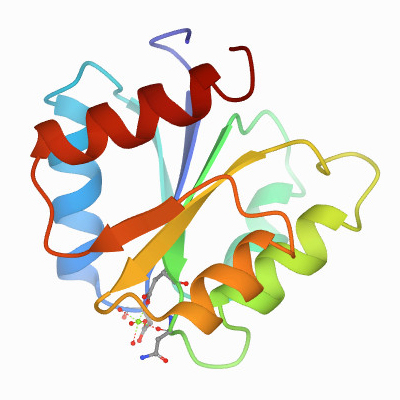
The first component turned out to be a kinase that phosphorylates itself on a histidine. Then the response regulator picks up the phosphoryl group from the histidine and transfers it to one of its own aspartates. This drives a conformational change within a regulatory domain that can be linked to almost any kind of effector domain. About two-thirds of them are DNA-binding domains and regulate transcription; some are enzymes, some are RNA-binding domains, some are protein–protein interaction domains. There’s no limit to what you can hook up to this phosphorylation-activated switch.
I wanted to learn structural biology in order to further the idea that these proteins were really similar. I collaborated with Greg Petsko and eventually was lucky enough to land a fellowship that required me to do a second postdoc, so I went to Greg’s lab. (Author’s note: Gregory Petsko is a professor at Harvard Medical School and a former president of the ASBMB.) When we determined the structure of the Salmonella chemotaxis protein CheY, it told the whole story . It was this small alpha–beta domain in which the three most highly conserved residues all clustered together. We knew immediately that this was the active site.
A lot of my career as an independent investigator has been trying to piece together what is conserved within two-component systems and what differs among them. We’ve learned that what is conserved is the enzymology: the core phosphoryl transfer and the high-energy aspartate phosphorylation that stabilizes what would otherwise be a less favorable conformation of the switch domain. Then any kind of regulation that exploits protein–protein interactions that discriminate between the two conformational states, whether activating or inhibiting interactions, is fair game for these proteins.
These days, are you taking a more computational approach?
Yes. Over 300,000 of these systems have been identified, and they do such diverse things. As people have done in vitro biochemistry with these proteins, it’s been found that their kinetic parameters differ by orders of magnitude. Some have a thousandfold more kinase or phosphatase activity than others. Similarly, their interactions can be orders of magnitude different in binding affinities.
There are also different ways that the components are configured. There are feedback loops in the transcription factors, where one of the operons that it regulates encodes itself. The question is, Why do you have autoregulation like this in some systems and not in others?
We’re trying to understand some of the design principles of the systems. Why do different systems have different kinetic parameters, different binding constants and different pathway architectures? What does that achieve in terms of the output response?
We’ve found that protein concentrations are really key. This shouldn’t surprise a biochemist. Autoregulation is designed to make just the right amount of protein at just the right time; if you start screwing around with that, the cells are not competitive. In studies of engineered strains with altered levels of proteins, within about 24 hours in chemostats, there’s so much selective pressure that mutations arise that convert the level back to the wild-type level. That tells you how important this regulation is.
A lot of the behavior of the pathway comes from having the right setting of the kinetic parameters and binding constants relative to the protein concentrations. More recently, we started wondering, “Well, what sets the protein concentrations?” and began work with a model system looking at transcription factors.
We started to ask whether the level of a transcription factor is at capacity for the number of binding sites in the cell. I pictured a high concentration of transcription factor and not a lot of binding sites. But it turns out that the transcription factors really are not numerically in great excess. By putting in additional binding sites on plasmids, we found that by just doubling the number of binding sites, you really change the output of the system.
It’s been very eye-opening. What we would consider minor perturbations in protein levels can have dramatic effects on the system. That should give us a little bit of caution in how we manipulate systems when we study them experimentally. For example, if a mutation affects the stability of the protein — and this happens frequently — our tendency is to attribute the alteration in system behavior to a functional consequence of the mutation that we’ve made rather than to throwing off the balance of protein concentrations.
Talk about your working relationship with research professor Rong Gao.
He’s been a wonderful partner. Rong has a real love of mathematics and computation and has been the driving force to bring us into this systems biology approach to understanding pathways. It’s a back-and-forth integration of mathematical modeling and experiments where the experimental data gives us enough parameters to begin the modeling; as soon as we get the model developed, we can make predictions from it and then go back and test it in the experimental system.
It’s really wonderful to have a senior person in the laboratory; first of all, to watch them develop, but also, as one progresses and becomes overburdened with many other activities, it can be incredibly useful to have someone who is still at the bench full time. I spent a big chunk of time in an administrative role. The founding director of our center passed away back in 2012. I took over as interim director, thinking that it would be a short-term thing, but ended up in this administrative position for almost eight years.
What did you learn from that leadership role?
I’ve learned from administration that I don’t want to do it. (Laughter.) I’m really enjoying being back in the lab.
So what made you decide to serve as ASBMB president?
I’ve hit a stage in my career that I’d like to be giving back a little bit. The ASBMB has meant a lot to me; I believe very strongly in the need for a collective voice for our discipline. And my involvement with the ASBMB has made me want to do more. I’ve met amazing people, and I’ve learned so much from them, and it’s given me a view of science from outside the narrow perspective of my own research.
What do you hope to achieve as president?
My agenda is to increase the engagement of members with the society. We are doing so much, but even amongst the most highly engaged members, people don’t know all the things that the ASBMB is doing. So my goal is to do as much as I can to broaden participation in committees and in activities of the society and let members find the activities that they feel most passionate about and get involved.
I understand that you have a twin sister who’s also in science. What’s that like?
It’s been a really strange trajectory. Growing up as identical twins, we were always trying to distinguish ourselves from each other. If she had long hair, I had short hair. If her favorite color was green, mine was blue. Then we went to college and lived on opposite sides of a giant school and never saw each other, and we found out that we’d bought many of the same clothes and albums. It was just too funny.
Our career story is almost like that. We both went to UC Berkeley because we had limited resources and tuition was free. My sister got into the business school and went down a career path in business, and I went down a science path. After graduating, she worked for a couple of years at Shell Oil and then obtained an MBA from Harvard Business School. She was hired by a consulting company where her clothing allowance when she signed was greater than my annual stipend. We were opposites.
Then she consulted on the health care sector and started working with pharma. She went on to Genzyme and got more and more into science, eventually becoming president of Genzyme Molecular Oncology. (Author’s note: Genzyme was a Boston-based biotechnology company that merged with Sanofi in 2011.) Since then, she has headed BayBio and several biotech firms in the San Franciso Bay area. She’s learned an enormous amount of science along the way. I mean, despite no formal training, she knows more immunology than I do.
Do you nerd out about molecules when you get together?
Not so much. In our early years, we used to argue, because I had a passion for basic science and thought decisions being made from a commercial standpoint were too profit-oriented. I’ve had postdocs apply to my lab after they’ve had their projects canned in pharma because overnight the company was no longer interested in what they were working on.
But although my passion is still for basic research, I have come to appreciate other perspectives. During an interview at Merck, my interviewer explained that there’s something really wonderful about knowing that what you do is going to impact people’s lives directly. And the things that are most valuable, dollarwise, are often the things that impact the most people in the biggest way. From that perspective, it makes sense that pharma chases dollars.
We’re meeting on the last day of the final Experimental Biology conference. What do you hope to see at the ASBMB’s first independent annual meeting next year?
I am eagerly looking forward to DiscoverBMB, our first independent meeting in many years, which will be held in Seattle in March next year. I am hoping to see much more networking and interaction among our community.
I think that with the small meeting format, we will have the ability to create a sense of community, including an emphasis on special interest groups. We’ll be trying to create a hub in the exhibit hall to bring people together, to showcase society activities, and to promote socialization and networking at strategic times throughout the meeting. We have a very enthusiastic pair of program planning committee co-chairs, Karen Allen and Craig Cameron, as well as a task force that is working diligently to develop plans for a vibrant and enjoyable meeting experience next year in Seattle.
One of the most important aspects of scientific conferences is the opportunity for networking and socialization among participants. I often find that informal conversations rather than structured presentations are the most valuable in sparking new ideas. The annual meeting is a great venue for young scientists to network with leaders in the field as well as for established scientist to reconnect with distant colleagues. We’ll be trying to promote such interactions in Seattle, and we’re looking for opportunities to celebrate science outside the walls of the convention center.

Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in People
People highlights or most popular articles
Sketching, scribbling and scicomm
Graduate student Ari Paiz describes how her love of science and art blend to make her an effective science communicator.

Embrace your neurodivergence and flourish in college
This guide offers practical advice on setting yourself up for success — learn how to leverage campus resources, work with professors and embrace your strengths.

Survival tools for a neurodivergent brain in academia
Working in academia is hard, and being neurodivergent makes it harder. Here are a few tools that may help, from a Ph.D. student with ADHD.

Quieting the static: Building inclusive STEM classrooms
Christin Monroe, an assistant professor of chemistry at Landmark College, offers practical tips to help educators make their classrooms more accessible to neurodivergent scientists.

Hidden strengths of an autistic scientist
Navigating the world of scientific research as an autistic scientist comes with unique challenges —microaggressions, communication hurdles and the constant pressure to conform to social norms, postbaccalaureate student Taylor Stolberg writes.
Richard Silverman to speak at ASBMB 2025
Richard Silverman and Melissa Moore are the featured speakers at the ASBMB annual meeting to be held April 12-15 in Chicago.

