
Predicting PROTAC properties
In recent decades, molecules that co-opt the ubiquitination system to degrade target proteins have opened up new drug targets by eliminating the need for an active site or a binding pocket. Proteolysis-targeting chimeras, or PROTACs, work by bringing together a drug target protein and a ubiquitin ligase. If it works as intended, PROTAC-induced proximity leads to target ubiquitinated and proteasomal degradation, removing the target from the cell. But sometimes the process stalls out.
“Sometimes binding can happen, but ubiquitination cannot happen,” said Nan Bai, a scientist at Amgen. Bai and colleagues sought to predict this undesirable outcome by modeling the many conformations that a multiprotein structure can adopt, in a workflow they reported in the Journal of Biological Chemistry.
PROTAC efficacy depends on two successful events. First, the drug must link its target protein with a ubiquitin ligase substrate receptor into what often is called a ternary complex; second, it must bring the ternary complex into a larger ubiquitin ligase holoenzyme where ubiquitin can be transferred onto a lysine in the target protein.
According to Bai, more effort has focused on predicting ternary complex formation, in part because it is easier. Her work focuses on the second step, triggering degradation. Her team used ensemble modeling to determine the most likely collection of conformations that a target PROTAC–ligase complex could adopt. They fit the group of structures that were most energetically favorable into the five most common conformations that a multiple-protein E3 ligase holoenzyme can adopt. In the resulting array of potential complexes, they searched for surface lysines eligible for ubiquitination on a target within reach of the holoenzyme. They scored each potential structure, deeming a complex unproductive if it predicted a steric clash or no lysines within the enzyme’s active zone. Then they predicted a compound’s overall ubiquitination efficiency based on the percentage of ensemble structures classed as productive.
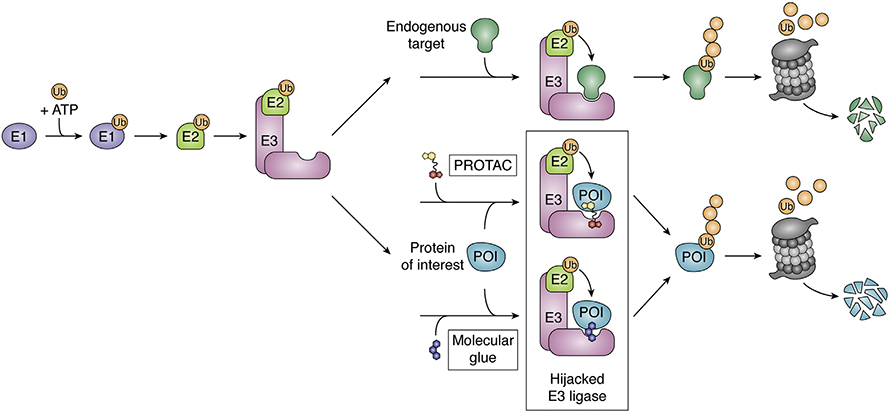
To validate this model, the researchers tested it on PROTAC–target pairs reported to form productive complexes with published structures and found good agreement. The model also suggested an explanation for perplexing previous results where a family of closely related kinases bound comparably to a PROTAC but showed dramatically different degradation rates. Among the poorly degraded targets, the team found a smaller proportion of possibly productive holoenzyme conformations. Collaborators at Promega conducted cellular assays of target degradation that further bolstered support for the model.
Sara Humphreys is a principal scientist at Amgen and senior author on the paper. “Before embarking on this work, a priori for an uncharacterized binding site on a protein, you really wouldn’t know” whether it would function as a PROTAC, she said.
While it provides useful information, Humphreys said, the model has not completely taken over the drug developers’ conversations about which molecules should advance in preclinical development. Although ubiquitination of a target is essential, it is not all there is to PROTAC efficacy; a target’s rate of synthesis, a complex’s proteasome recruitment, and the dynamics of ubiquitin chain elongation and deubiquitination can all affect whether a target is destroyed. In the future, other tools may tackle these variables.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Opinions
Opinions highlights or most popular articles

Making my spicy brain work for me
Researcher Reid Blanchett reflects on her journey navigating mental health struggles through graduate school. She found a new path in bioinformatics, proving that science can be flexible, forgiving and full of second chances.

The tortoise wins: How slowing down saved my Ph.D.
Graduate student Amy Bounds reflects on how slowing down in the lab not only improved her relationship with work but also made her a more productive scientist.
How pediatric cataracts shaped my scientific journey
Undergraduate student Grace Jones shares how she transformed her childhood cataract diagnosis into a scientific purpose. She explores how biochemistry can bring a clearer vision to others, and how personal history can shape discovery.

Debugging my code and teaching with ChatGPT
AI tools like ChatGPT have changed the way an assistant professor teaches and does research. But, he asserts that real growth still comes from struggle, and educators must help students use AI wisely — as scaffolds, not shortcuts.

AI in the lab: The power of smarter questions
An assistant professor discusses AI's evolution from a buzzword to a trusted research partner. It helps streamline reviews, troubleshoot code, save time and spark ideas, but its success relies on combining AI with expertise and critical thinking.

How AlphaFold transformed my classroom into a research lab
A high school science teacher reflects on how AI-integrated technologies help her students ponder realistic research questions with hands-on learning.

