Toward ‘harmonizing’ lipidomics
Scientists have been studying lipids and their effects on the human body for many decades. As new technologies emerge and yield great volumes of data, those who work in the emerging field of lipidomics are under increasing pressure to ensure reproducibility and greater transparency. A new paper in the Journal of Lipid Research represents another step toward those goals, reporting the results of a lipidomics comparison study performed by 31 labs in the U.S. and abroad.
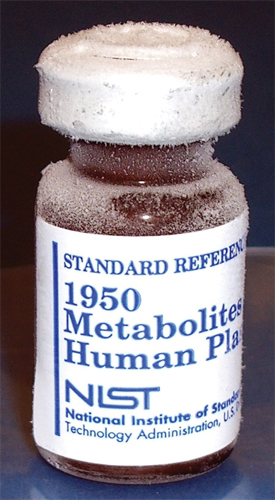 Participants in the study used their own methods and conventions to measure lipids in this reference material, SRM 1950.Courtesy of NIST
Participants in the study used their own methods and conventions to measure lipids in this reference material, SRM 1950.Courtesy of NIST  John Bowden
John Bowden
The project, led by John Bowden of the National Institute of Standards and Technology, required all participating laboratories to use their own methods and conventions to measure lipids in NIST’s Standard Reference Material 1950. The team picked SRM 1950 (“Metabolites in Frozen Human Plasma”) because it is commercially available and was made from plasma of 100 individuals who are representative of the U.S. population.
In the end, the cohort reported more than 1,500 unique lipids (arranged by sum composition), and NIST’s stats team determined consensus location estimates and uncertainties (probable concentration and range) for 339 lipids that were reported by at least five laboratories.
ASBMB Today’s executive editor, Angela Hopp, talked to Bowden about what spurred him to do the interlaboratory comparison exercise and how it may influence lipidomics as the field continues to mature. The interview has been edited for length and clarity.
What motivated you to undertake this study?
Everybody is employing different work flows, and the technology to measure lipids is rapidly advancing. This is combined with the fact that more and more people are getting into the field and the fact that maybe those people who get into the field don’t know the impact of measurement decisions or don’t have any experience with how to actually process data.
NIST is interested in making an impact on the measurement science of lipids. But we first need to establish a baseline for how lipids are currently measured across the community. We (asked): “How much variation exists in the community right now with all the different methodologies and philosophies noted for measuring lipids?”
We gave all the laboratories the SRMs, frozen and homogenous materials, and we said: “Measure the lipids quantitatively using your routine lipidomic methodologies. Measure what you’re comfortable measuring. Provide us the data. And then, using that data, we can assess the weaknesses and strengths in how things are measured and what is the extent of the variation that exists within the community.
We can now begin to figure out how to continue improving measurement practices and get to the next step, which is communitywide harmonization. From there, we can then begin discussing efforts toward standardization.
How can people use the information your project generated?
The NIST Statistical Engineering Department generated several consensus mean estimates with uncertainties for lipid data submitted by all laboratories. Consensus means estimates signify the probable concentration and range, using the uncertainty, for a lipid measured in the SRM.
If you want to extend quality-control activities from the intralaboratory level to interlaboratory level, there are not many tools to do this type of activity. So, by providing these consensus means for specific SRMs, labs can now buy that material and they can assess how they’re doing in comparison to a good cross-section of the community.
You know, the kind of stuff we’re doing is not sexy but it is important. It can be hard sometimes to get people excited about the measurement science of lipids.
If lipidomics is going to continue to expand and mature, in the clinical field or any field, there have to be best-practice guidelines. There have to be metrics to assess data quality. Of course, those things don’t come overnight. You have to get the community interested in efforts aimed at improving harmonization, and I think we have.
You sense that this is becoming more valued by the community.
We had about a 30 percent participation rate for this study. (Author’s note: Bowden invited 100 labs to participate.) Well, one of the questions on our follow-up survey was “Have you participated in an intercomparison study?” And a bunch of people said they had, (but) not a lot. And there was another question: “Would you like to participate in a (future) interlab study?” And we had, I think, over 100 people say “yes.” I think that that is because, over the last three years, I’ve been presenting at conferences. I’ve been networking. We’ve been spreading the message of metrology.
I think when this work gets published and people start embracing harmonization, you’re eventually going to start seeing journals requiring authors to provide more details about their lipidomics data before they publish articles. With metabolomics right now, there are certain guidelines — certain things you have to address or you have to mention in your paper. You know, if you go to get grants, hopefully there’s going to be a way for people to assess whether you are a laboratory that provides quality measurements or not.
There are very few ways to assess the quality of someone’s lipidomics data. In my survey, I think over 90 percent of the people don’t put their data in repositories. This needs to change; we need to promote more transparency with the data we collect and publish.
In the paper, you mentioned that this is all a bit polarizing. How so?
So, it’s polarizing on multiple levels. If you talk about how things are measured, in terms of methodology, there are different camps. For example, targeted versus untargeted lipidomics.
It’s also polarizing when you get into the finer details of measurement. So, for instance, if you’re talking about quantitation. There’s a lot of different philosophies on how you’re supposed to quantitate, even down to the point of how you define your quantitation — like whether it’s absolute, relative, semi, etc. How you define lipid quantitation, just the definition of that — people have different philosophies.
For example, in most fields quantitation requires a standard measured at multiple concentrations, which almost exactly matches your compounds of interest. In untargeted lipidomics, a standard with slightly different properties at only one concentration may be used. Some people still refer to this method as quantitation; some say relative quantitation; some say semiquantitation. But we don’t have a consensus on what words to use. These words are important; they let you know whether the concentration reported is a rough estimate or can be considered accurate within a defined level of uncertainty.
As you dig deeper into the fundamentals of how lipids are measured, more and more things come to light, and everybody in the community is going to have their own philosophy on how to address each aspect of measurement.
We have some follow-up manuscripts (coming), and one of them is actually taking method information, whether it be their extraction method or how they’re processing data or their mass spec approach, and looking at how different methodology drives certain trends in the submitted data.
Have you had any pushback?
I think most researchers recognize the need for better measurement efforts and best-practice guidelines. Even those who did not participate in the exercise, many of them indicated that they supported the efforts. I think the resistance, if any, will come when best-practice guidelines are introduced, as I’m sure people will have differing opinions on what the guidelines should entail.
I think, ultimately, that the community will have to realize that we all need to work together.
I’m not waiting for it to happen. I’ll keep pushing.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
CRISPR epigenome editor offers potential neurodevelopmental gene therapies
Scientists from the University of California, Berkeley, created a system to modify the methylation patterns in neurons. They presented their findings at ASBMB 2025.

Finding a symphony among complex molecules
MOSAIC scholar Stanna Dorn uses total synthesis to recreate rare bacterial natural products with potential therapeutic applications.

E-cigarettes drive irreversible lung damage via free radicals
E-cigarettes are often thought to be safer because they lack many of the carcinogens found in tobacco cigarettes. However, scientists recently found that exposure to e-cigarette vapor can cause severe, irreversible lung damage.

Using DNA barcodes to capture local biodiversity
Undergraduate at the University of California, Santa Barbara, leads citizen science initiative to engage the public in DNA barcoding to catalog local biodiversity, fostering community involvement in science.

Targeting Toxoplasma parasites and their protein accomplices
Researchers identify that a Toxoplasma gondii enzyme drives parasite's survival. Read more about this recent study from the Journal of Lipid Research.

Scavenger protein receptor aids the transport of lipoproteins
Scientists elucidated how two major splice variants of scavenger receptors affect cellular localization in endothelial cells. Read more about this recent study from the Journal of Lipid Research.

