New kids on the block: Base and prime editors
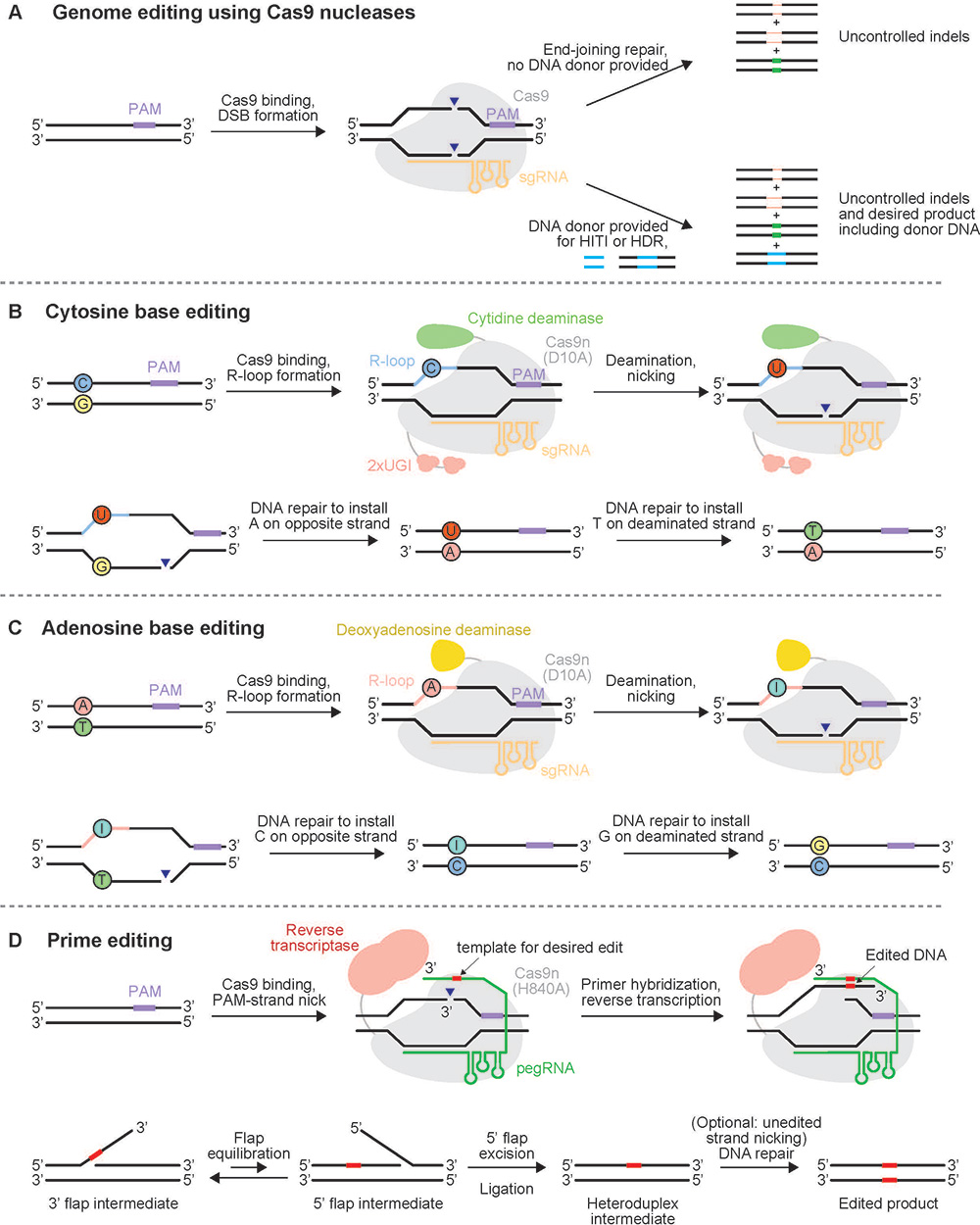
CRISPR–Cas9 was first described in 2012 as “genetic scissors.” Even though this system has revolutionized research, medicine, agriculture and biotechnology, it is not without its weaknesses. CRISPR–Cas9 induces double-strand DNA breaks in the genome. These breaks are usually repaired by nonhomologous end joining machinery, which is prone to creating insertions, deletions or translocations and could cause undesired or even unsafe side effects.
DNA breaks made by CRISPR-Cas9 are well suited to disrupt or delete a targeted portion of the genome, which is the mechanism of the first-ever Food and Drug Administration-approved gene therapy for sickle cell disease in December. However, researchers say their low efficiency can make developing them for medical applications challenging.
To address these pitfalls, David Liu and colleagues developed two novel genetic engineering methods that build upon CRISPR–Cas9.
As described in the late 2010s, base and prime editors are more efficient than the CRISPR–Cas9 system. Instead of inducing double-strand breaks, base and prime editors take advantage of a catalytically impaired Cas9, known as Cas9 nickase, which cuts only one strand of the DNA at the target location.
“CRISPR–Cas9 is generally more genotoxic than base and prime editors,” said Greg Newby, assistant professor of genetic medicine at Johns Hopkins School of Medicine and former Liu lab member. “If genotoxicity leads to an increased rate of cancer or a less effective therapy, base or prime editing could end up being a better therapeutic approach.”
Two spinoff companies from the lab, Beam Therapeutics and prime medicine, are pursuing base and prime editing approaches to cure genetic diseases. Beam Therapeutics is conducting ongoing clinical trials for hemoglobinopathies, such as sickle cell disease and beta thalassemia.

Base editors are composed of a DNA targeting domain like Cas9 nickase fused to a cytidine or adenosine deaminase and a targeting guide RNA. This complex of molecular machines mediates a cytosine to thymine substitution in the target genomic DNA. Though the editing capability of base editors is limited, they are extremely efficient, Newby said.

Conversely, prime editors are made up of a Cas9 nickase fused to a reverse transcriptase and a prime editing guide RNA. According to Liu, these editors offer a “search and replace genome editing method” that can make a wide variety of changes to a sequence with minimal off-target effects.
“Unlike base editing, you can tell the prime editor exactly what change you want to make whether that’s a small insertion, deletion, any point mutation or combination of the three,” Kelcee Everette, a graduate student at Harvard University and member of the Liu lab, said.
Newby, Everette and colleagues used base and prime editors to develop gene therapies for sickle cell disease. In a series of recent studies, they demonstrated that both base and prime editing methods could rescue sickle cell disease in mice.
A 2021 study from Newby and colleagues showed that a custom base editor could convert the sickle cell disease mutant gene to a nonpathogenic, rare variant known as hemoglobin G Makassar, which generally has the properties of normal adult hemoglobin. After performing base editing of hematopoietic stem and progenitor cells outside of the host and reintroducing them back into a mouse model of sickle cell disease, 68% of beta globin in the blood had been successfully edited and remained stable over more than 16 weeks. In addition, red blood cell sickling and splenic inflammation were reduced.
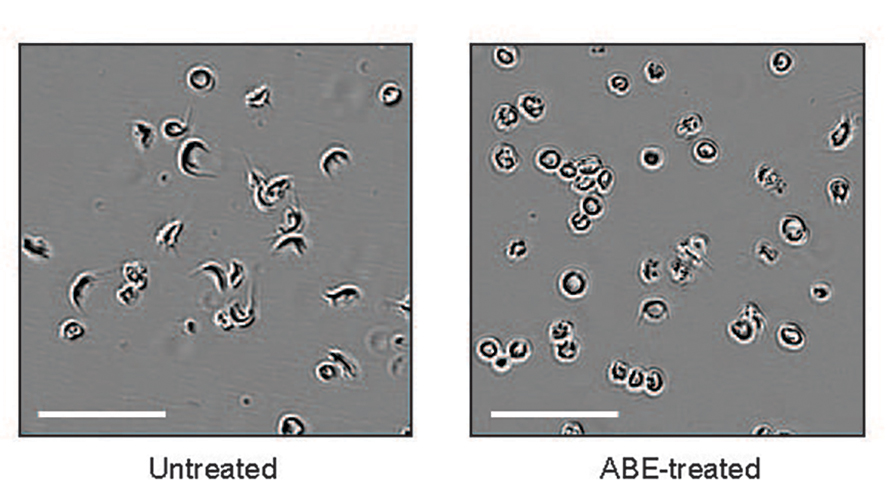
Similarly, Everette and colleagues showed in a 2023 study that a prime editor could correct the sickle cell mutant to the normal sequence. Everette prime-edited human stem and progenitor cells from sickle cell disease patients and transplanted them into immunodeficient mice. After 17 weeks, 42% of the erythrocytes, or red blood cell precursors, showed the prime-edited sequence, demonstrating that this approach is durable.
Everette said that this prime editing approach may not be as efficient as base editing, but it induces fewer off-target mutations and therefore may turn out to be safer after more optimization.
Though both approaches from Newby and Everette’s studies would require myeloablation and a hematopoietic stem cell transplant in humans, they and their colleague André Lieber are researching delivery methods, such as adenoviral vectors and lipid nanoparticles. These methods would directly deliver base and prime editors to their target cells in the human body without the need for myeloablation or a stem cell transplant.
“I hope that this kind of therapy will be available in the long run so that patients do not have to undergo a painful bone marrow transplant,” Newby said.
Learn more
Read about the development of the CRISPR-Cas9 therapy for sickle cell disease that was recently approved by the U.S. Food and Drug Administration in "The Road to Survival."
Enjoy reading ASBMB Today?
Become a member to receive the print edition monthly and the digital edition weekly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Genetics studies have a diversity problem that researchers struggle to fix
Researchers in South Carolina are trying to build a DNA database to better understand how genetics affects health risks. But they’re struggling to recruit enough Black participants.
Scientists identify new function of learning and memory gene common to all mammalian brain cells
Findings in mice may steer search for therapies to treat brain developmental disorders in children with SYNGAP1 gene mutations.
From the journals: JBC
Biased agonism of an immune receptor. A profile of missense mutations. Cartilage affects tissue aging. Read about these recent papers.
Cows offer clues to treat human infertility
Decoding the bovine reproductive cycle may help increase the success of human IVF treatments.
Immune cells can adapt to invading pathogens
A team of bioengineers studies how T cells decide whether to fight now or prepare for the next battle.
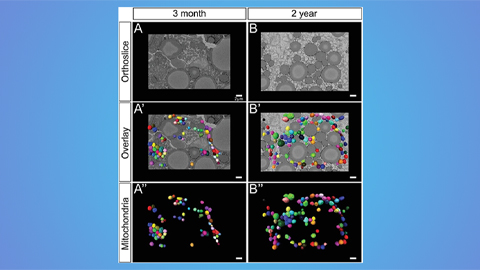
Hinton lab maps structure of mitochondria at different life stages
An international team determines the differences in the 3D morphology of mitochondria and cristae, their inner membrane folds, in brown adipose tissue.

